Gallery: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 242: | Line 242: | ||
util.cbaw | util.cbaw | ||
bg white | bg white | ||
set light_count, | set light_count,8 | ||
set spec_count,1 | set spec_count,1 | ||
set shininess, 10 | set shininess, 10 | ||
| Line 434: | Line 434: | ||
# soften out the image | # soften out the image | ||
set light_count, | set light_count,8 | ||
set spec_count,1 | set spec_count,1 | ||
set shininess, 10 | set shininess, 10 | ||
Revision as of 15:35, 28 February 2013
| Cool PyMOL-generated Images and their Scripts. Add Your Own |
| Complex B&W outline representation | What To Type | |||||
|
# first load lipid model
load lipids.pdb;
# hide the initially loaded representation
hide all;
# set background color to white
bg_color white;
# show lipid model as sticks
show sticks, lipids;
# color the lipids model by element CHNOS #2 (carbon green)
util.cbag lipids;
# select all hydrogens and remove them from the model
select hideme, hydro;
hide everything, hideme;
delete hideme;
# create phosphate spheres
create phos, elem p;
hide everything, phos;
show spheres, phos;
# load helix model
load helix.pdb;
# hide the initially loaded representation
hide everything, helix;
# make the helical struct into a cartoon form
show cartoon, helix;
# style the cartoon form
cartoon putty;
# reposition the helix among the lipids using
# the 3-Button Editing Mouse Mode
# basically
# Shift+Left Mouse to rotate the helix
# Shift+Middle Mouse to move the helix
# also, you may want to make liberal use of the
# get_view and set_view commands.
#
# When you have the scene set like you want,
# continue with...
# move the model to find the view you want,
# and use get_view to get the coordinate description
get_view;
# set ray_trace_mode to black and white outline
set ray_trace_mode, 2;
|
| Grid Mode | What To Type | |||||
|
fetch 1cll 1sra 1ggz 5pnt 1rlw 1cdy;
set grid_mode
Hint: You may wish to execute the 'reset' command on the command line after running the above commands to get full molecules in view of window and centered in a more useable manner. |
| Cool Perspective | What To Type | |||||
|
load prot.pdb;
zoom i. 46-49 and n. CA
set field_of_view, 60
ray
|
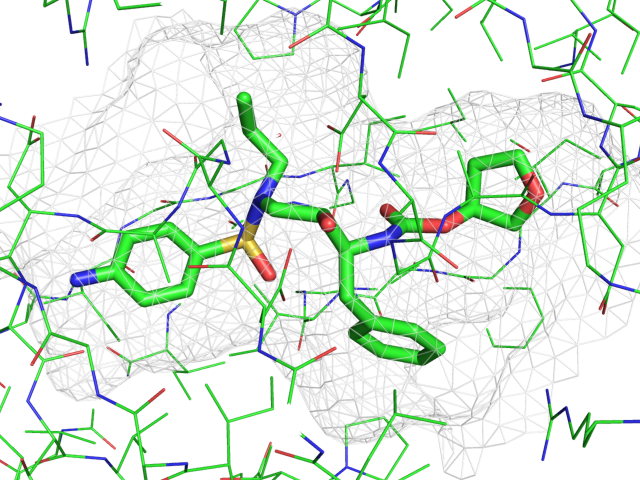
| Representing a binding pocket | What To Type | |||||
|
load $TUT/1hpv.pdb, tmp
extract lig, organic
extract prot, polymer
delete tmp
set surface_carve_cutoff, 4.5
set surface_carve_selection, lig
set surface_carve_normal_cutoff, -0.1
show surface, prot within 8 of lig
set two_sided_lighting
set transparency, 0.5
show sticks, lig
orient lig
set surface_color, white
set surface_type, 2 # mesh
unset ray_shadows
|
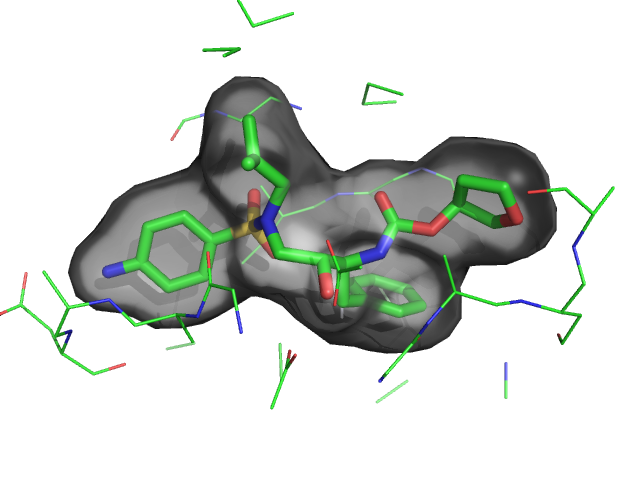
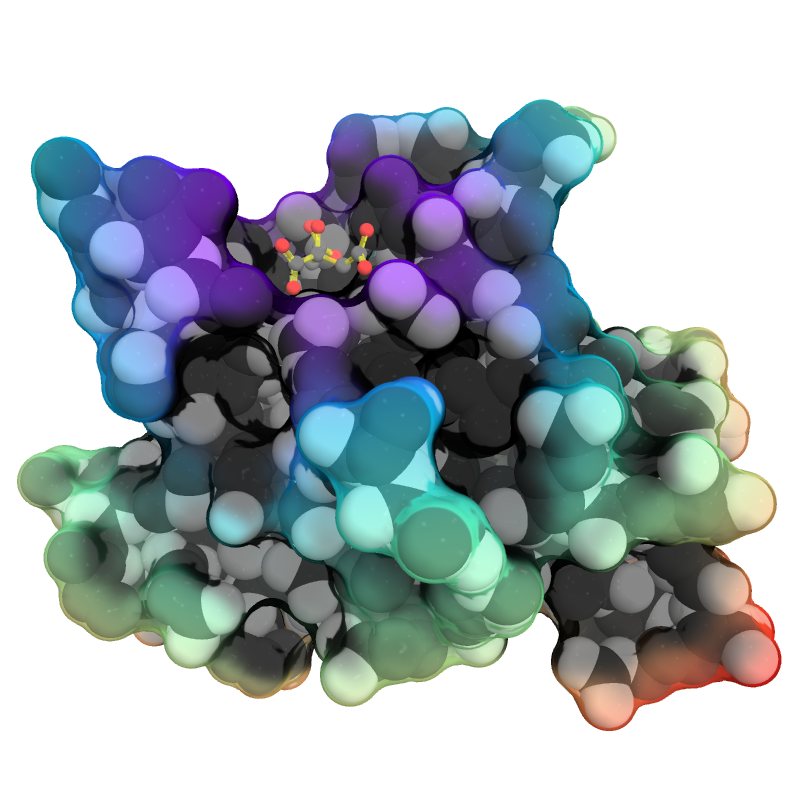
| QuteMol Like | What To Type | |||||
|
load $TUT/1hpv.pdb
set_color oxygen, [1.0,0.4,0.4]
set_color nitrogen, [0.5,0.5,1.0]
remove solvent
as spheres
util.cbaw
bg white
set light_count,8
set spec_count,1
set shininess, 10
set specular, 0.25
set ambient,0
set direct,0
set reflect,1.5
set ray_shadow_decay_factor, 0.1
set ray_shadow_decay_range, 2
unset depth_cue
# for added coolness
# set field_of_view, 60
ray
|
| Simulating Tilt-shift | What To Type | |||||
|
fetch 1wld
as surface, poly
as sticks, org
h_add solvent
color grey, poly
orient org
png img.png
# now, go into Photoshop or the GIMP and apply a Gaussian or
# Focus blur to the top and bottom portions of the image
|
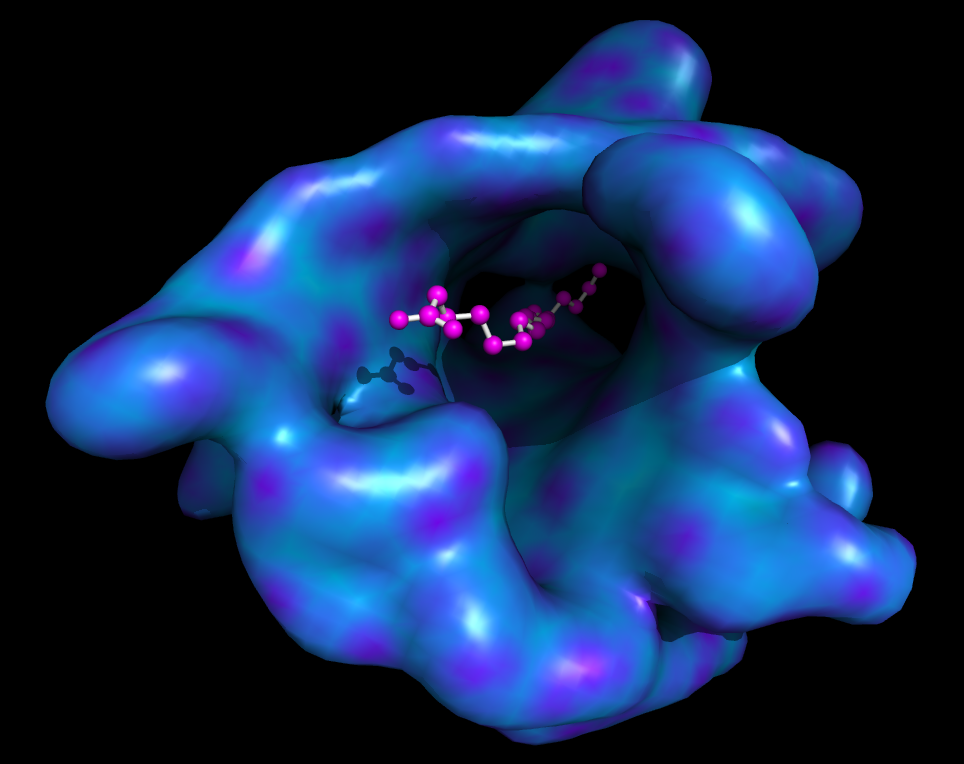
| Ray-normal-based transparency | What To Type | |||||
|
# grey surface
set surface_color, grey
# cavity mode
set surface_mode, 3
# layered transparency mode
set transparency_mode, 1
# surface transparency
set transparency, 0.5
# oblique and contrast define the
# look of the surface transparency:
# if the normal vector is
set ray_transparency_oblique
set ray_transparency_oblique_power, 8
set ray_transparency_contrast, 7
# fetch a protein, with a
# small molecule in a nice
# hidden pocket
fetch 1hpv, async=0
hide
# show the small molecule as surface
show surface, org
# arrange the view
orient org
# zoom back a little
zoom org, 1
# show the small molecule inside as sticks
show sticks, org
# show some nearby sidechains
show lines, poly within 5 of org
# enable frame caching for playback
set cache_frames, 1
set ray_trace_frames, 1
mset 1x120
movie.roll 1, 120, 1, x
mplay
# now sit back and wait 5 minutes...
|
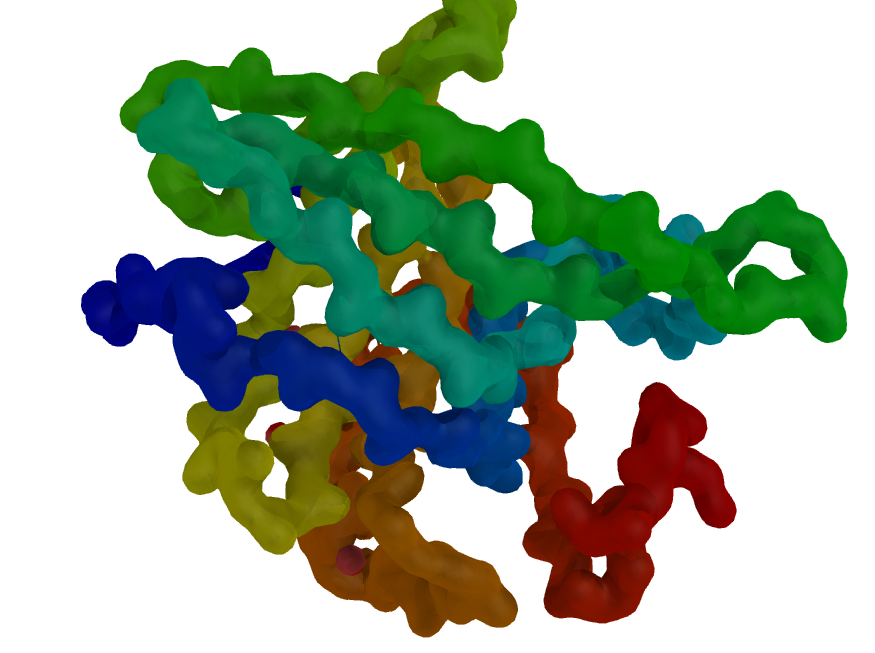
| Blobby Main Chain | What To Type | |||||
|
fetch 3uex, struct, async=0
remove solvent
# set the B-factors nice and high for smoothness
alter all, b=10
alter all, q=1
# 3.5 A map resolution
set gaussian_resolution, 3.5
# new gaussian map w/resolution=0.5 Ang
# on just the main chain
map_new map, gaussian, 0.5, n. C+O+N+CA, 5
# create a surface from the map
isosurface surf, map, 3.0
# color the protein by number
spectrum count, rainbow, struct
# now color the map based on the underlying protein
cmd.ramp_new("ramp", "struct", [0,10,10], [-1, -1, 0])
# set the surface color
cmd.set("surface_color", "ramp", "surf")
# hide the ramp and lines
disable ramp
hide lines
bg grey
# soften out the image
set light_count,8
set spec_count,1
set shininess, 10
set specular, 0.075
set ambient,0
set direct,0
set reflect, 0.85
set ray_shadow_decay_factor, 0.1
set ray_shadow_decay_range, 4
unset depth_cue
# ray trace the image
orient
ray
|
| Blobby Side Chains | What To Type | |||||
|
# fetch a protein
fetch 1rx1, async=0
# setup the cartoon tubes
as cartoon
cartoon tube
set cartoon_tube_radius, 0.7
set cartoon_color, brown
set cartoon_side_chain_helper, on
show sticks, poly
color yellow
# README
# stop here, or try this for "sloppy sticks"
# beefy video card required!
select rep sticks
select sele and not n. C+O+N+CA
# set the B-factors nice and high for smoothness
alter all, b=10
alter all, q=1
# 2.5 A map resolution
set gaussian_resolution, 2.5
# 0.2 A sampling; lower=smoother
map_new map, gaussian, 0.2, sele, 5
# create a surface from the map
isosurface surf, map, 5.0
color yellow, surf
hide sticks
# reconnect the main chain to the blobs
show sticks, n. CA+CB
|
| Smooth surface with ligand | What To Type | |||||
|
fetch 3uex, struct, async=0
remove solvent
# set the B-factors nice and high for smoothness
alter all, b=10
alter all, q=1
# 3.5 A map resolution
set gaussian_resolution, 7.6
# new gaussian map w/resolution=0.5 Ang
# on just the main chain
map_new map, gaussian, 1, n. C+O+N+CA, 5
# create a surface from the map
isosurface surf, map, 1.5
# color the protein by number
spectrum count, rainbow, struct
# now color the map based on the b-factors of the
# underlying protein
cmd.ramp_new("ramp", "struct", [0,10,10], "rainbow")
# set the surface color
cmd.set("surface_color", "ramp", "surf")
# hide the ramp and lines
disable ramp
hide lines
show sticks, org
show spheres, org
color magenta, org
reinit
fetch 3uex, struct, async=0
remove solvent
# set the B-factors nice and high for smoothness
alter all, b=10
alter all, q=1
# 3.5 A map resolution
set gaussian_resolution, 7.6
# new gaussian map w/resolution=0.5 Ang
# on just the main chain
map_new map, gaussian, 1, n. C+O+N+CA, 5
# create a surface from the map
isosurface surf, map, 1.5
# color the protein by number
spectrum count, rainbow, struct
# now color the map based on the b-factors of the
# underlying protein
cmd.ramp_new("ramp", "struct", [0,10,10], "rainbow")
# set the surface color
cmd.set("surface_color", "ramp", "surf")
# hide the ramp and lines
disable ramp
hide lines
show sticks, org
show spheres, org
color magenta, org
set_bond stick_radius, 0.13, org
set sphere_scale, 0.26, org
set_bond stick_radius, 0.13, org
set_bond stick_color, white, org
set sphere_scale, 0.26, org
set_view (\
-0.877680123, 0.456324875, -0.146428943,\
0.149618521, -0.029365506, -0.988305628,\
-0.455291569, -0.889327347, -0.042500813,\
-0.000035629, 0.000030629, -37.112102509,\
-3.300258160, 6.586110592, 22.637466431,\
8.231912613, 65.999290466, -50.000000000 )
# ray trace the image
ray
|
| Complex Stylized Protein | What To Type | |||||
|
fetch 1eaz, async=0
extract oo, org
hide everything, solvent
set field_of_view, 50
preset.ball_and_stick("oo")
set_bond stick_color, 0xffff44, oo
set_bond stick_transparency, 0.35, oo
color grey, oo and e. C
set valence, 1, oo
ramp_new pRamp, oo, selection=poly, range=[5,30], color=rainbow
set surface_color, pRamp, poly
show spheres, poly
color white, poly
color grey30, poly and e. C
set sphere_scale, 0.99, poly
set ray_transparency_contrast, 0.20
set ray_transparency_oblique, 1.0
set ray_transparency_oblique_power, 20
show surface, poly
set surface_quality, 2
set light_count, 5
set ambient_occlusion_mode, 1
set ambient_occlusion_scale, 50
set ambient, 0.40
set transparency, 0.50
disable pRamp
set spec_power, 1200
set spec_reflect, 0.20
set ray_opaque_background, 0
set ray_shadow, 0
ray
|
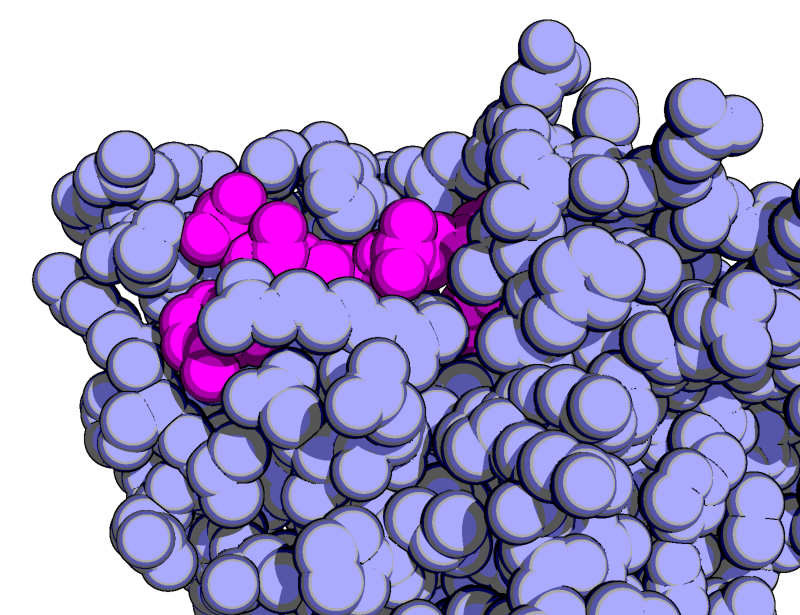
| Goodsell-like | What To Type | |||||
|
# fetch the protein
fetch 1rx1, async=0
# show it as blue/magenta spheres
as spheres
color lightblue, not org
color magenta, org
remove solvent
# set the view
orient all within 8 of org
# set the lights, ray tracing setttings
# to get the Goodsell-like rendering
unset specular
set ray_trace_gain, 0
set ray_trace_mode, 3
bg_color white
set ray_trace_color, black
unset depth_cue
ray
|