Gallery: Difference between revisions
Jump to navigation
Jump to search
(added image to gallery) |
No edit summary |
||
| Line 10: | Line 10: | ||
{{GalleryImage | {{GalleryImage | ||
<!-- This creates a row entry for the Gallery section --> | <!-- This creates a row entry for the Gallery section --> | ||
|<!-- Image --> | |<!-- Image --> | ||
image=BW_raytraced_complex_image.jpg|size=200px | image=BW_raytraced_complex_image.jpg|size=200px | ||
|<!-- Title above image --> | |<!-- Title above image --> | ||
title=Complex B&W outline representation | title=Complex B&W outline representation | ||
|<!-- 'Description' section ( | |<!-- 'Description' section (shows up below image)--> | ||
description=Making a B&W outlined image with depth. | description=Making a B&W outlined image with depth. | ||
|<!-- 'What to Type' section --> | |<!-- 'What to Type' section --> | ||
| Line 89: | Line 85: | ||
<li>Some parts of 'Layer 1' are transparent and shouldn't be. Using the Paint Bucket tool fill in these areas with white (or whichever color you find appropriate).</li> | <li>Some parts of 'Layer 1' are transparent and shouldn't be. Using the Paint Bucket tool fill in these areas with white (or whichever color you find appropriate).</li> | ||
<li>Admire your handiwork; put it in a publication, presentation, or poster.</li></ol> | <li>Admire your handiwork; put it in a publication, presentation, or poster.</li></ol> | ||
|<!-- 'See Also' section --> | |<!-- 'See Also' section (shows up below image)--> | ||
seeAlso= | seeAlso= | ||
* [[Bg_Color|bg_color]] | * [[Bg_Color|bg_color]] | ||
Revision as of 10:12, 26 August 2008
| Cool PyMOL-generated Images and their Scripts. Add Your Own |
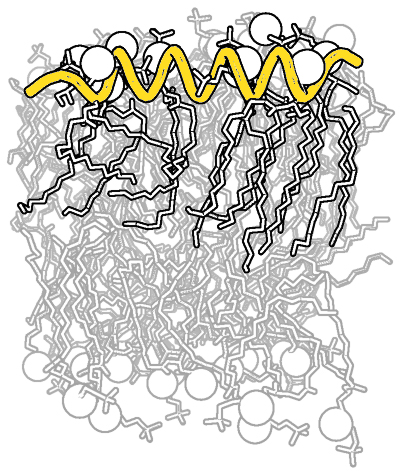
| Complex B&W outline representation | What To Type | |||||
|
# first load lipid model
load lipids.pdb;
# hide the initially loaded representation
hide all;
# set background color to white
bg_color white;
# show lipid model as sticks
show sticks, lipids;
# color the lipids model by element CHNOS #2 (carbon green)
util.cbag lipids;
# select all hydrogens and remove them from the model
select hideme, hydro;
hide everything, hideme;
delete hideme;
# create phosphate spheres
create phos, elem p;
hide everything, phos;
show spheres, phos;
# load helix model
load helix.pdb;
# hide the initially loaded representation
hide everything, helix;
# make the helical struct into a cartoon form
show cartoon, helix;
# style the cartoon form
cartoon putty;
# reposition the helix among the lipids using
# the 3-Button Editing Mouse Mode
# basically
# Shift+Left Mouse to rotate the helix
# Shift+Middle Mouse to move the helix
# also, you may want to make liberal use of the
# get_view and set_view commands.
#
# When you have the scene set like you want,
# continue with...
# move the model to find the view you want,
# and use get_view to get the coordinate description
get_view;
# set ray_trace_mode to black and white outline
set ray_trace_mode, 2;
|
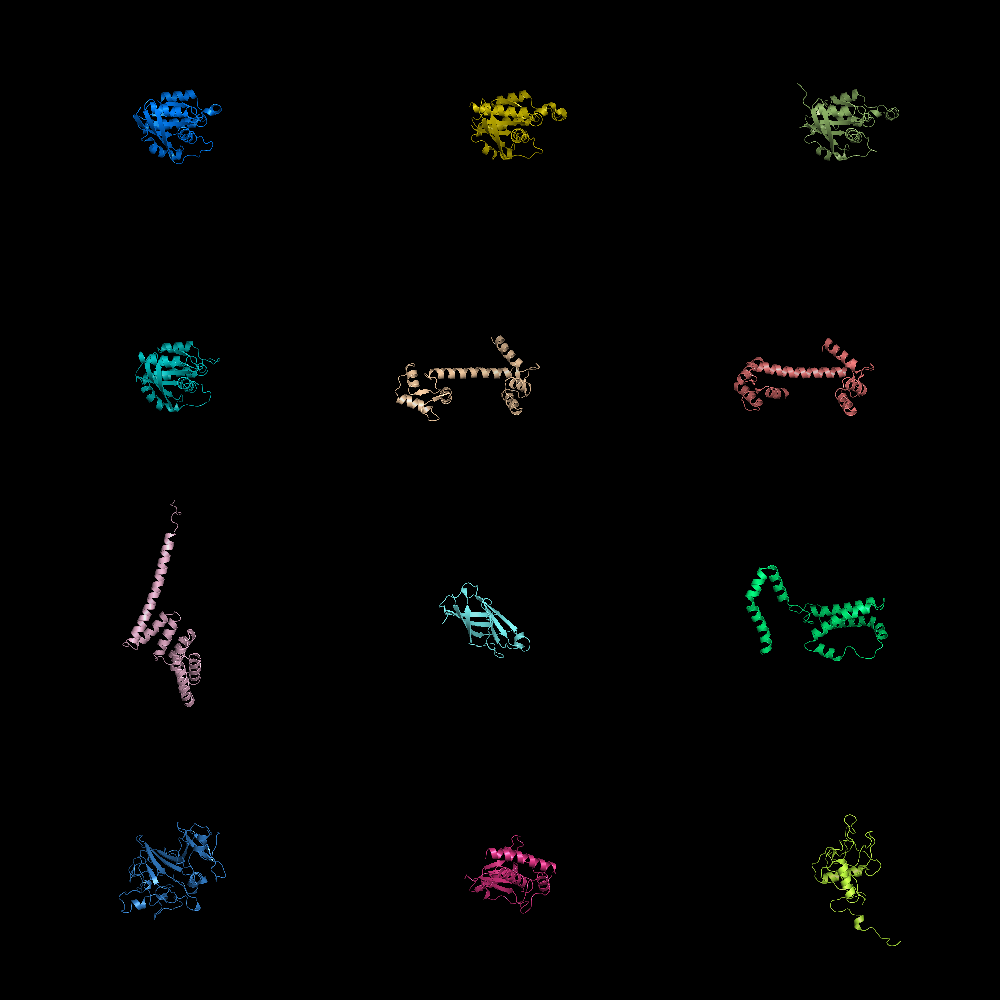
| Grid Mode | What To Type | |||||
|
fetch 1cll 1sra 1ggz 5pnt 1rlw 1cdy;
set grid_mode
|
| Cool Perspective | What To Type | |||||
|
load prot.pdb;
zoom i. 46-49 and n. CA
set field_of_view, 60
ray
|
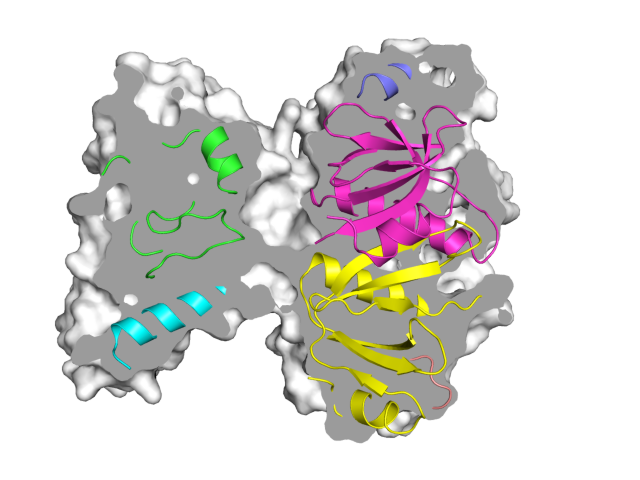
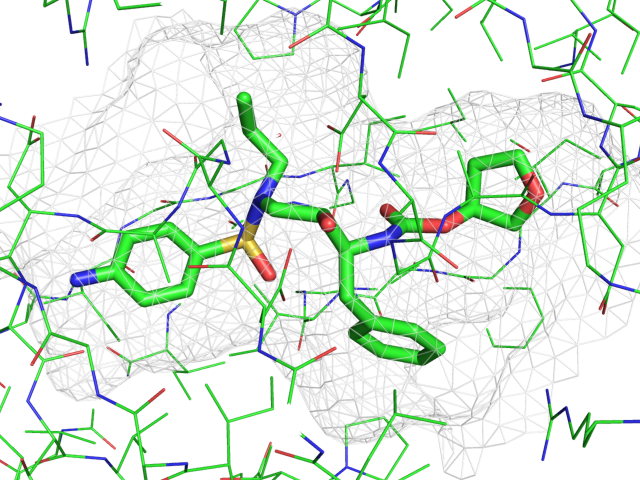
| Representing a binding pocket | What To Type | |||||
|
load $TUT/1hpv.pdb, tmp
extract lig, organic
extract prot, polymer
delete tmp
set surface_carve_cutoff, 4.5
set surface_carve_selection, lig
set surface_carve_normal_cutoff, -0.1
show surface, prot within 8 of lig
set two_sided_lighting
set transparency, 0.5
show sticks, lig
orient lig
set surface_color, white
set surface_type, 2 # mesh
unset ray_shadows
|
| QuteMol Like | What To Type | |||||
|
load $TUT/1hpv.pdb
set_color oxygen, [1.0,0.4,0.4]
set_color nitrogen, [0.5,0.5,1.0]
remove solvent
as spheres
util.cbaw
bg white
set light_count,10
set spec_count,1
set shininess, 10
set specular, 0.25
set ambient,0
set direct,0
set reflect,1.5
set ray_shadow_decay_factor, 0.1
set ray_shadow_decay_range, 2
unset depth_cue
# for added coolness
# set field_of_view, 60
ray
|