Surface carve selection: Difference between revisions
Jump to navigation
Jump to search
(better examples) |
(gif) |
||
| Line 1: | Line 1: | ||
[[File:surface_carve_selection.gif||thumb|right|Animation with a moving pseudoatom as the surface_carve_selection]] | |||
With the '''surface_carve_selection''' and '''surface_carve_cutoff''' settings, the visibility of a molecular [[surface]] can be limited to the proximity of an atom selection. | With the '''surface_carve_selection''' and '''surface_carve_cutoff''' settings, the visibility of a molecular [[surface]] can be limited to the proximity of an atom selection. | ||
Latest revision as of 16:22, 24 February 2017
With the surface_carve_selection and surface_carve_cutoff settings, the visibility of a molecular surface can be limited to the proximity of an atom selection.
Unlike when showing surface representation only for a subset of atoms, the visibility is controlled on the vertex (surface point) level, not the atom level.
The value of surface_carve_selection must be a named selection (created with select), it cannot be a selection expression (e.g. "chain A" won't work).
Example
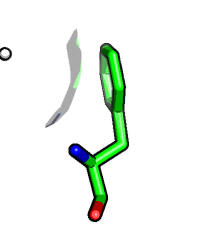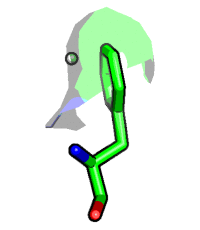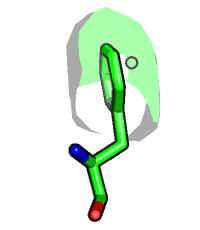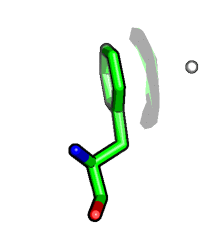
Visibility control on the vertex level. Hide surface points which are more than 5 Angstrom away from carvesele:
fetch 1rx1, async=0 as cartoon select carvesele, organic set surface_carve_selection, carvesele set surface_carve_cutoff, 5 show surface show sticks, carvesele
Compare to visibility control on the atom level. Hide surface for atoms which are more than 5 Angstrom away from carvesele.
fetch 1rx1, async=0 as cartoon select carvesele, organic show surface, all within 5 of carvesele show sticks, carvesele