Uploads by Jaredsampson
Jump to navigation
Jump to search
This special page shows all uploaded files.
| Date | Name | Thumbnail | Size | Description | Versions |
|---|---|---|---|---|---|
| 21:26, 13 February 2022 | Cgo examples.jpg (file) | 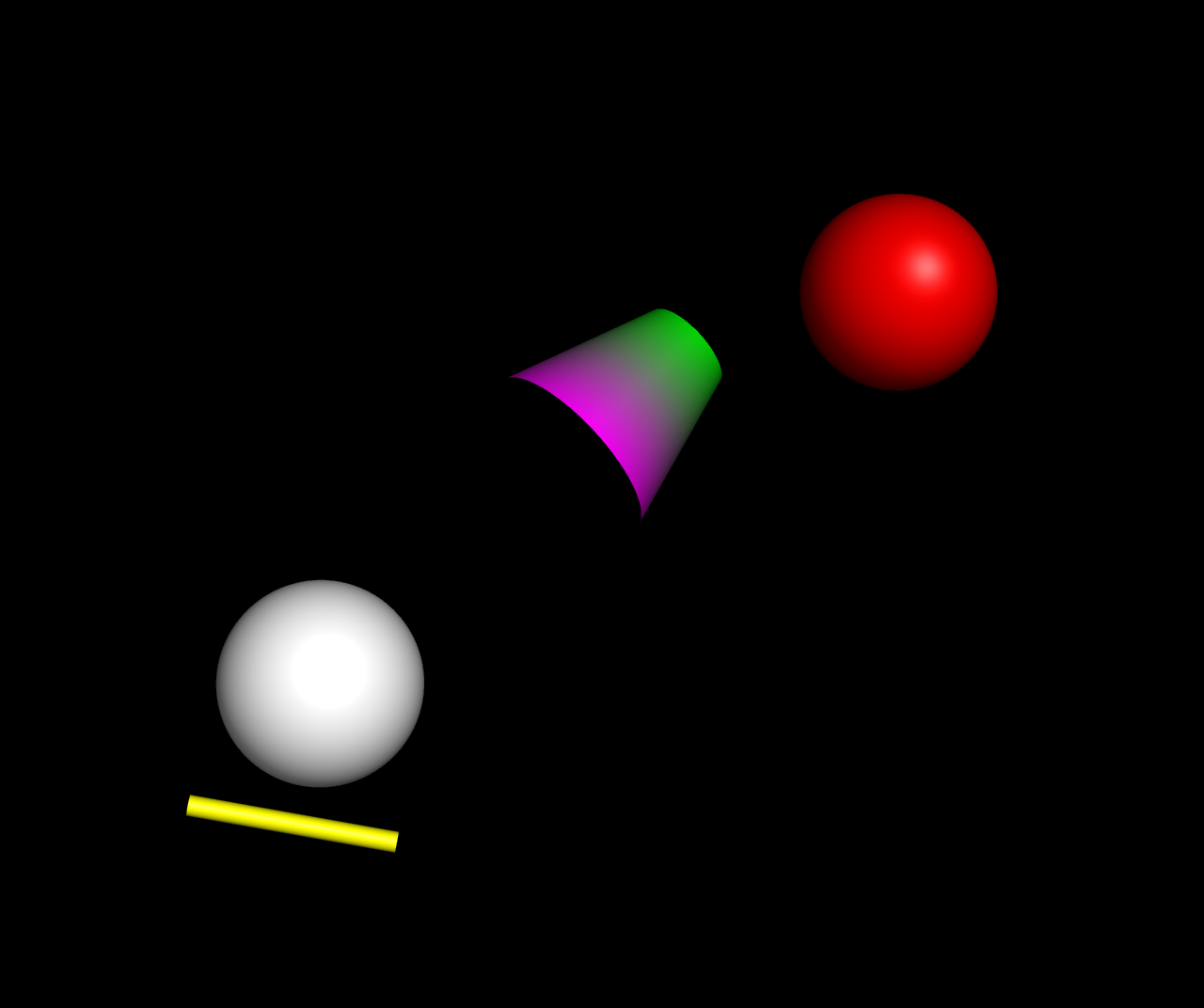 |
63 KB | Assorted rendered CGO objects in PyMOL. | 1 |
| 23:04, 5 December 2018 | 4gms colors.png (file) |  |
289 KB | Prepared in response to a pymol-users thread from 11/30/2018-12/5/2018 about the appearance of transparency in a published figure. Script to reproduce: bg_color white fetch 4gms, async=0 # hemagglutinin selections select ha, chain A select l130, ha... | 1 |
| 23:17, 25 October 2017 | Colorblindfriendly menu.png (file) | 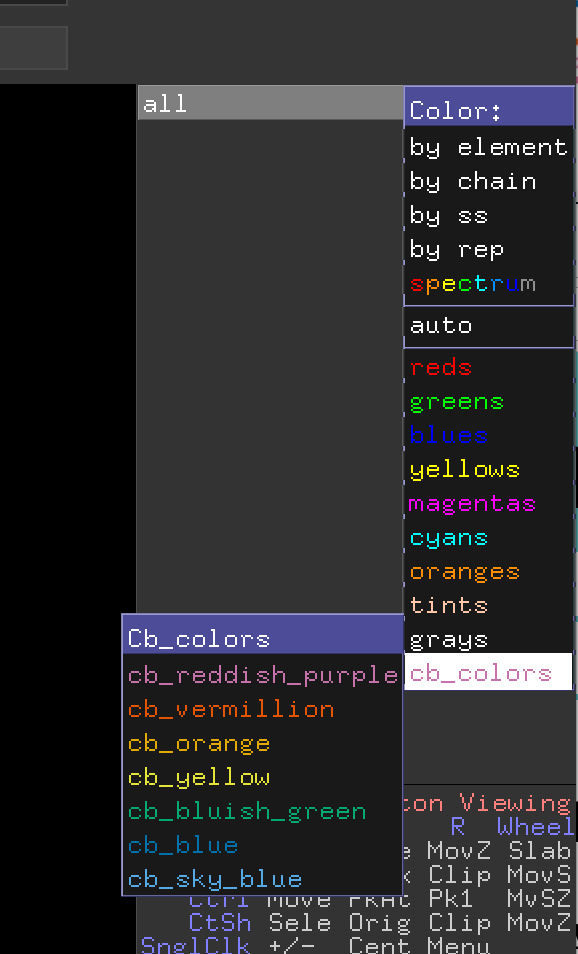 |
42 KB | Screenshot of the color list menu added to the OpenGL GUI by the colorblindfriendly.py script in PyMOL 2.0. | 1 |
| 12:39, 14 January 2017 | PyMOLProbity GUI.png (file) | 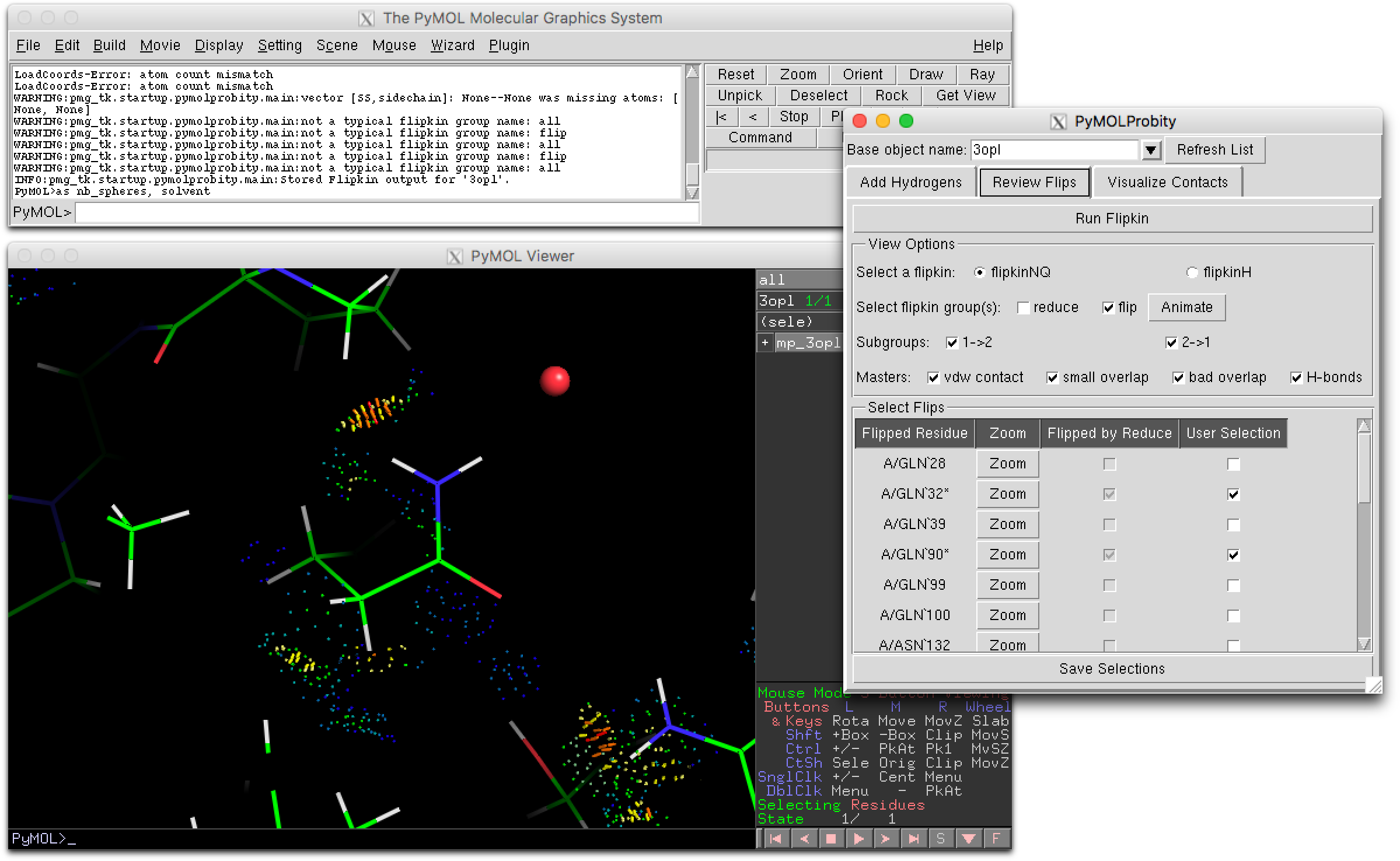 |
211 KB | Screenshot of the "Review Flips" tab of the PyMOLProbity plugin GUI, where flippable side chains in the selected object can be inspected, animated between unflipped (i.e. Reduce-output) and flipped orientations, and selected for future operations. | 1 |
| 11:47, 24 July 2014 | Outline cleaned.png (file) | 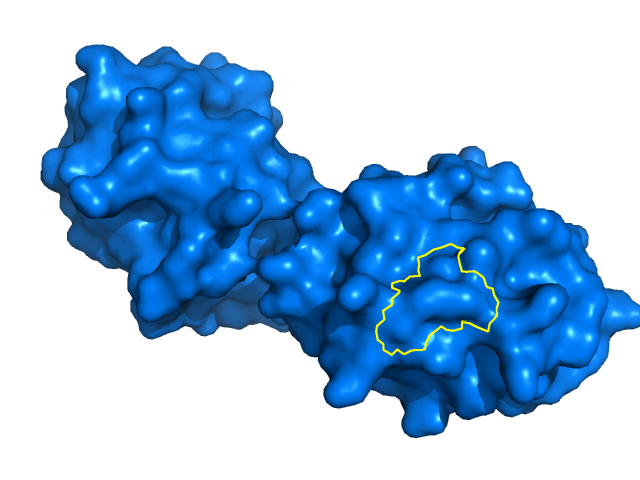 |
168 KB | Cleaned-up version of Outline_composite.png, with extra outline lines erased. | 1 |
| 11:45, 24 July 2014 | Outline composite.png (file) | 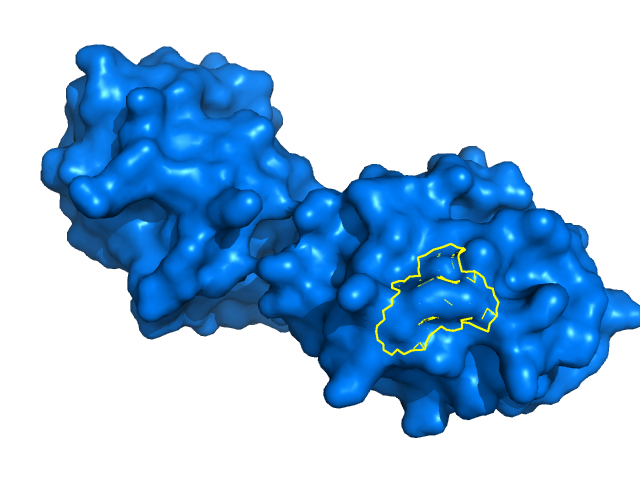 |
170 KB | Composite of Outine_base.png and Outline_overlay.png. | 1 |
| 11:42, 24 July 2014 | Outline overlay.png (file) |  |
7 KB | Cytochrome C (3cyt) residues within 2Å of Lys85 are outlined in yellow, using `ray_trace_mode` and `ray_trace_color` settings. | 1 |
| 11:39, 24 July 2014 | Outline base.png (file) |  |
166 KB | Surface representation of Cytochrome C (3cyt). | 1 |
| 10:56, 26 April 2013 | 20091111 Structure cover.jpg (file) |  |
130 KB | Cover of the 11 November 2009 issue of Structure. Artwork by Jared Sampson, Xiangpeng Kong lab, New York University School of Medicine. | 1 |
| 13:45, 8 June 2012 | Stanfield.png (file) | 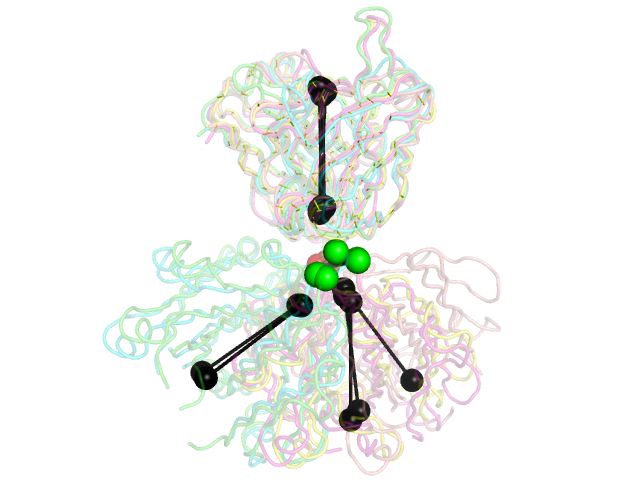 |
199 KB | Example PDBs from Stanfield, et al., JMB 2006 as shown in Fig. 1 of that paper. elbow_angle.py was used to calculate the elbow angles and draw a representation of the vectors used in the calculation. | 1 |
| 13:29, 8 June 2012 | Elbow angle 3ghe.png (file) | 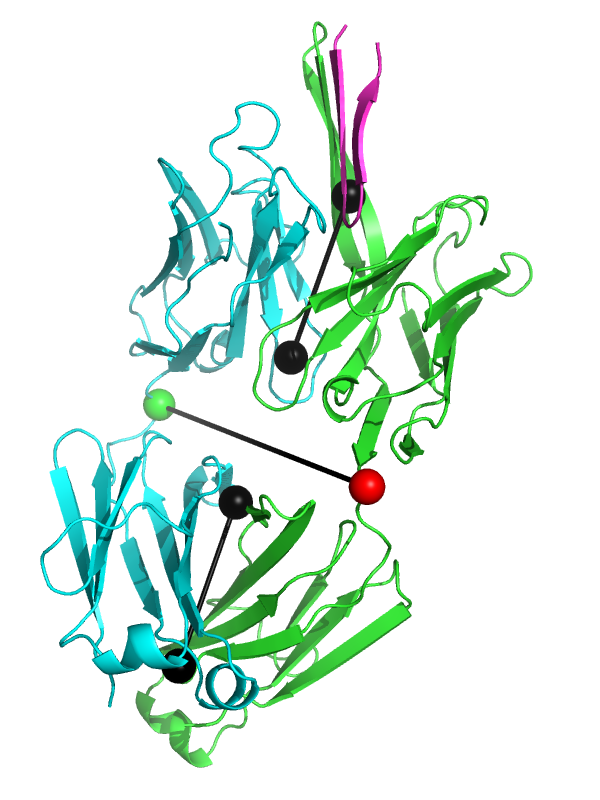 |
191 KB | View of the elbow_angle vector representation for the Fab fragment from PDB 3ghe. | 1 |
| 12:29, 7 September 2010 | 1hiw orient2.png (file) | 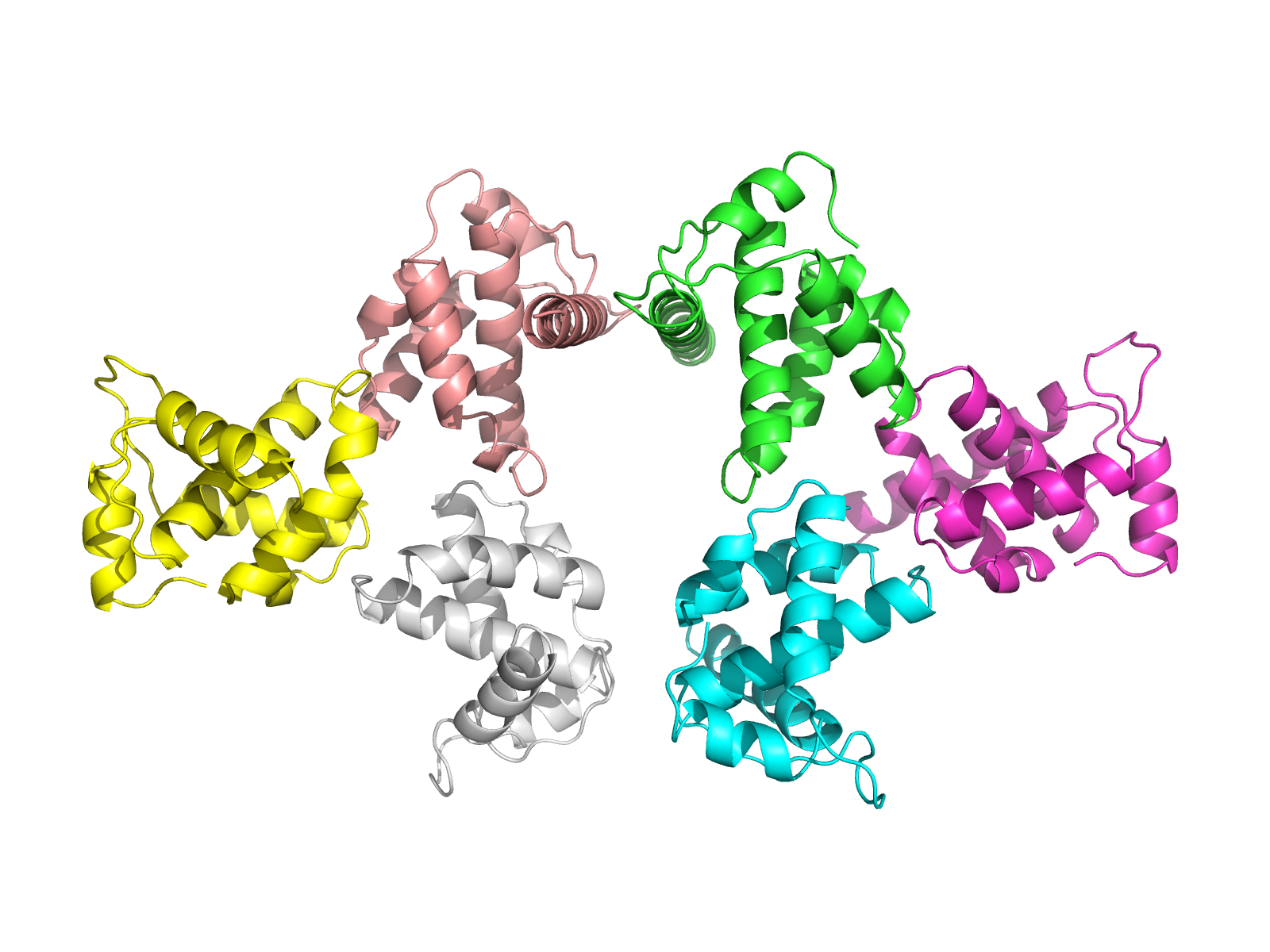 |
567 KB | Demonstration of orient command with 1hiw, two trimers shown. | 1 |
| 12:15, 7 September 2010 | 1hiw orient.png (file) | 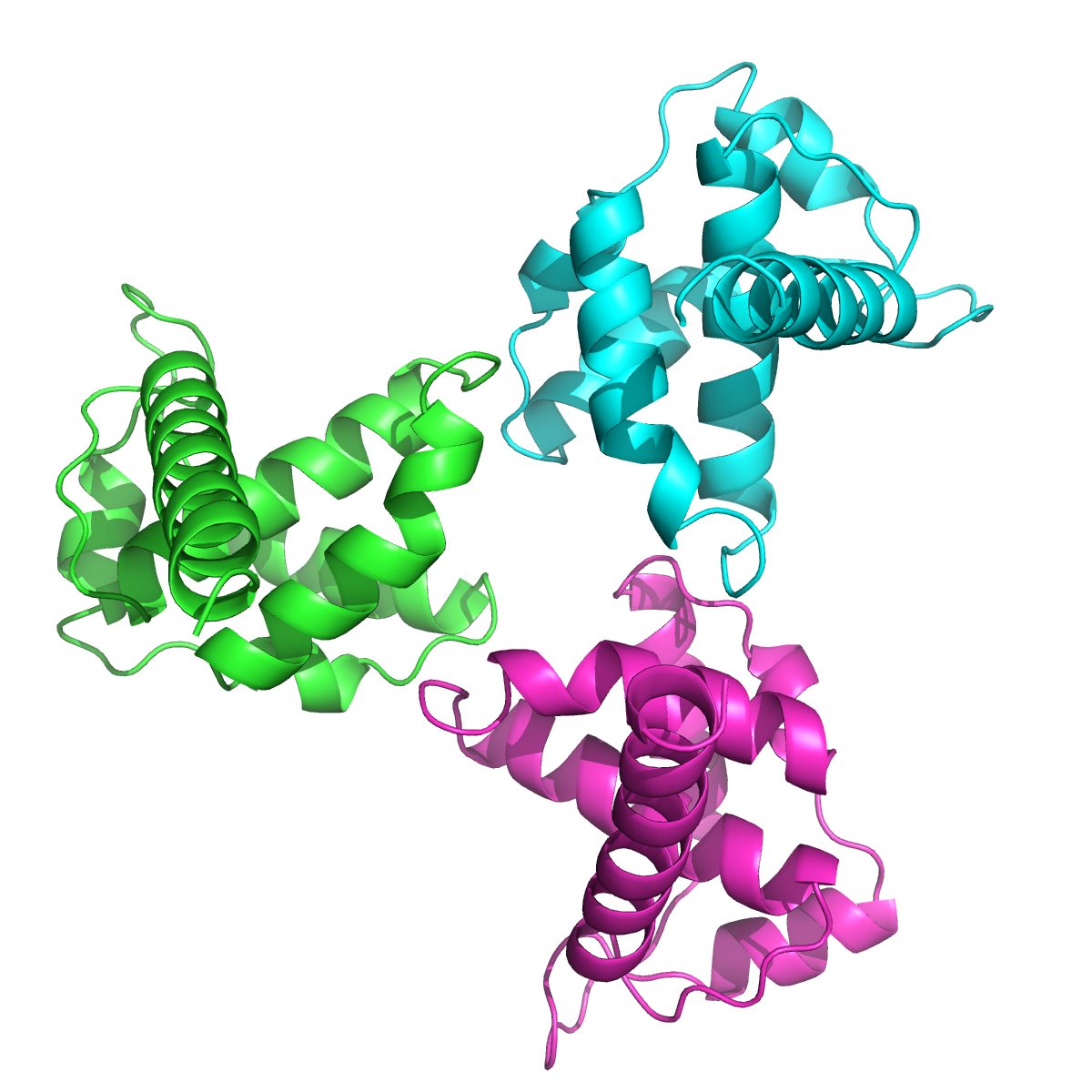 |
484 KB | Demonstration of orient command with NCS symmetry of PDB 1hiw. | 1 |