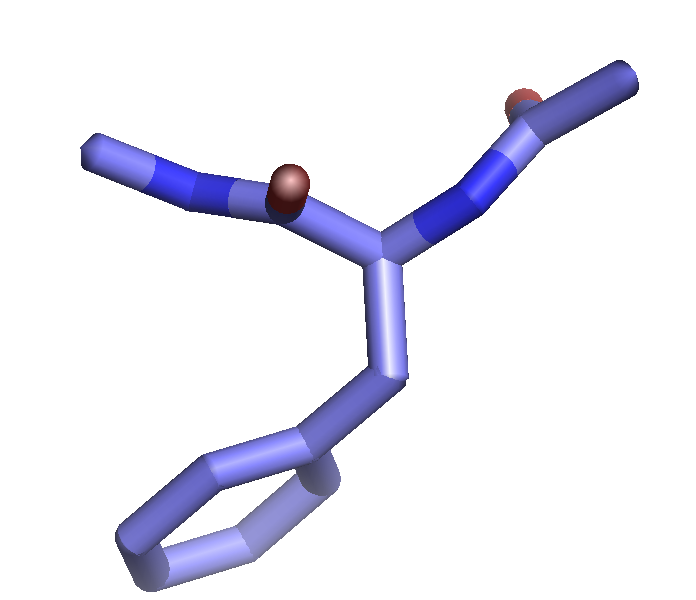
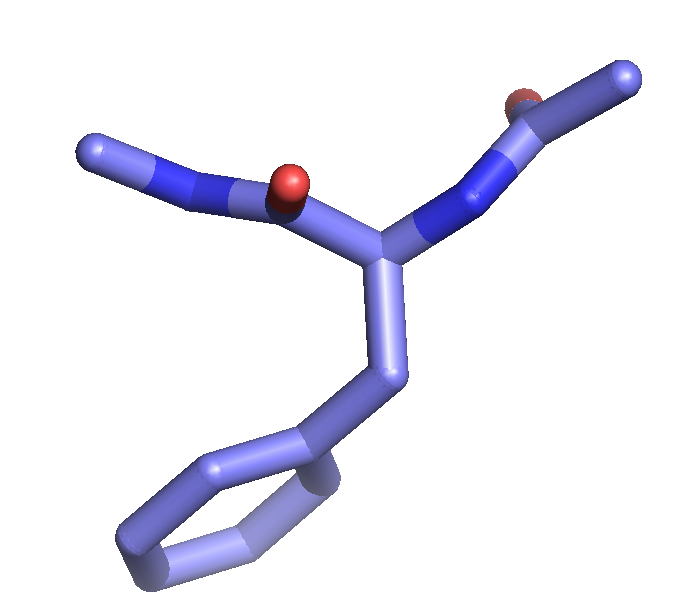
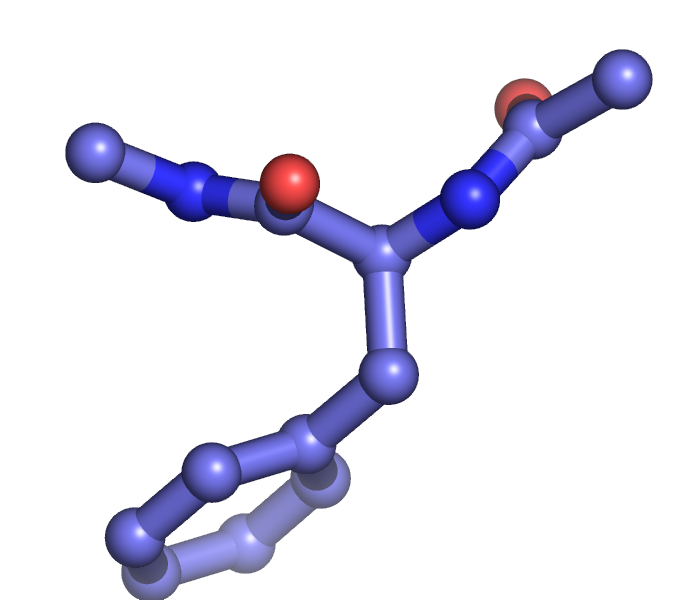
The setting "stick_ball_ratio" controls the relative ratio between the radius of sticks connecting bonded atoms and the radius of the atom spheres. Note that simply setting "stick_ball, on" will result in balls with the same radius as the sticks and so will appear only slightly different (the joins will be smoother). Changing the stick_ball_ratio without setting "stick_ball, on" will -- obviously -- have no apparent effect.
set stick_ball_ratio, 1.5
set stick_ball, on # displays atoms as balls joined by sticks
set stick_ball, off # displays only connected sticks