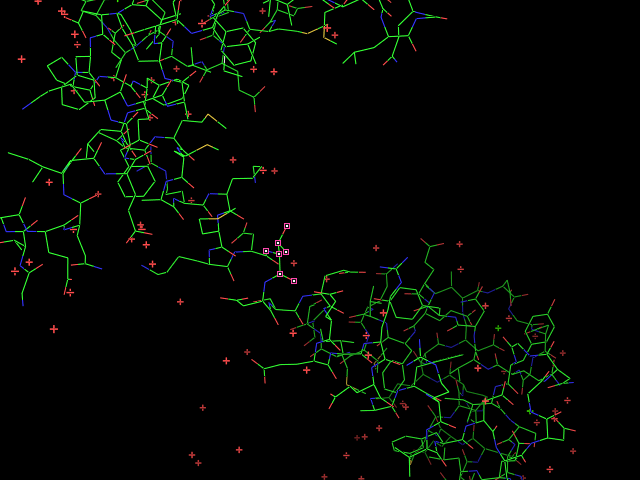
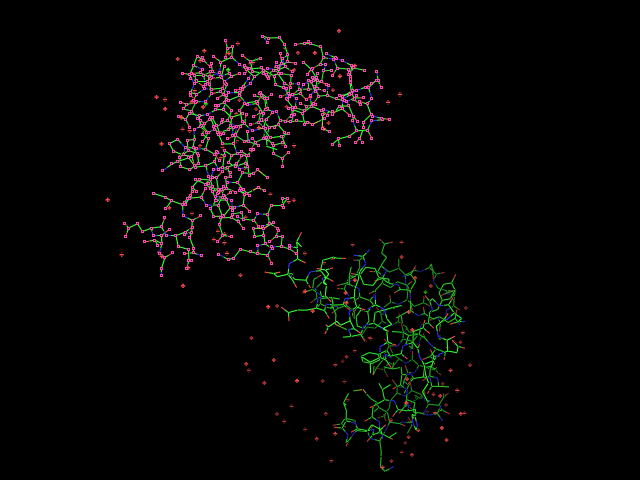
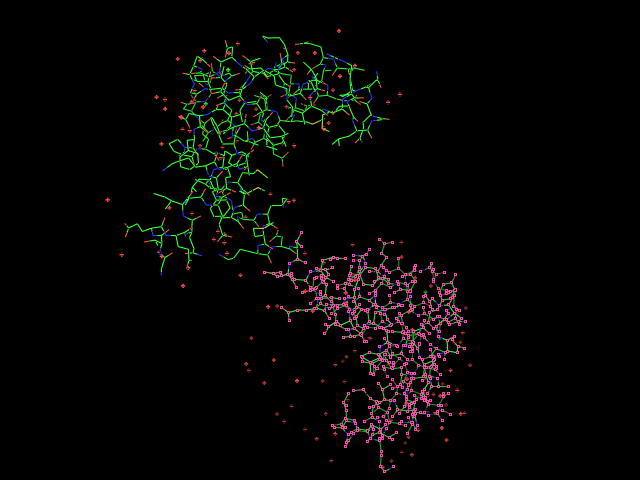
Given an initial selection,
Split_selection will create two new selections. One, called 'lo,' will have all the atoms with IDs lower than your input atom (actually input residue's alpha carbon); and the second selection is 'hi,' which contains the atoms with IDs higher than the initial (residue's alpha carbon) atom.
The Code
<source lang="python">
import pymol
from pymol import cmd
def get_index_list(s):
"""
Given an atom selection, return the list of atom IDs in this selection
"""
return map(lambda x: x.index, cmd.get_model(s).atom)
def get_single_index(s):
"""
Get the ID of the first alpha carbon in the selection, S
"""
# assume CA
return get_index_list( "n. CA and (br. %s)" % s)[0]
def split_selection(s):
"""
PARAMS
s
An atom selection.
RETURNS
None
SIDE EFFECTS
Creates two new selections, called lo and hi. Lo will have ..→