Distance
Jump to navigation
Jump to search
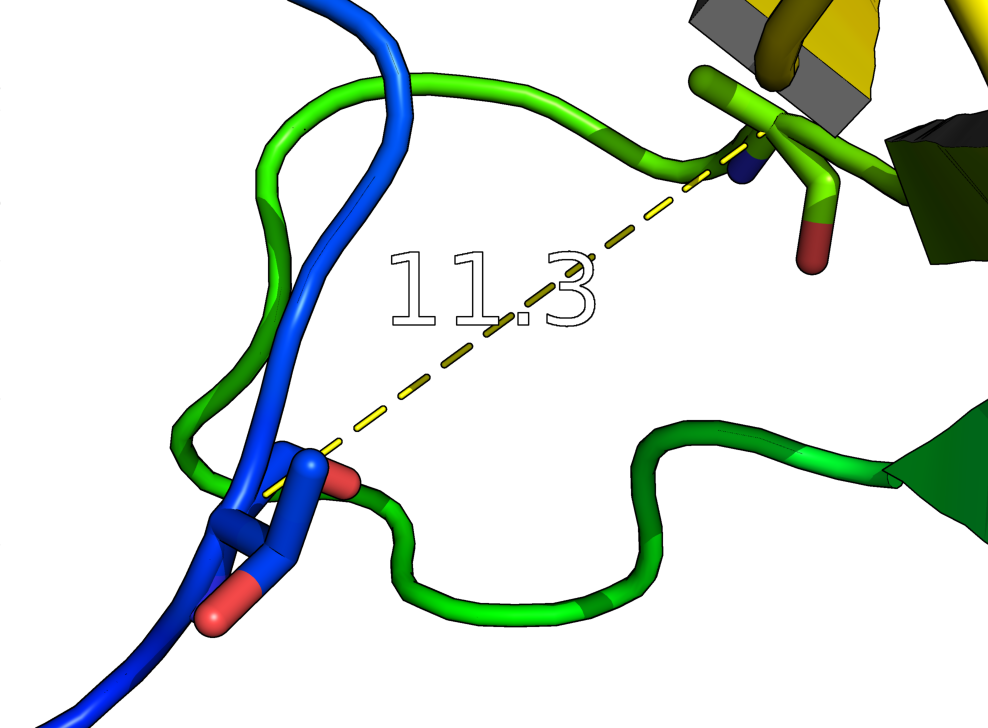
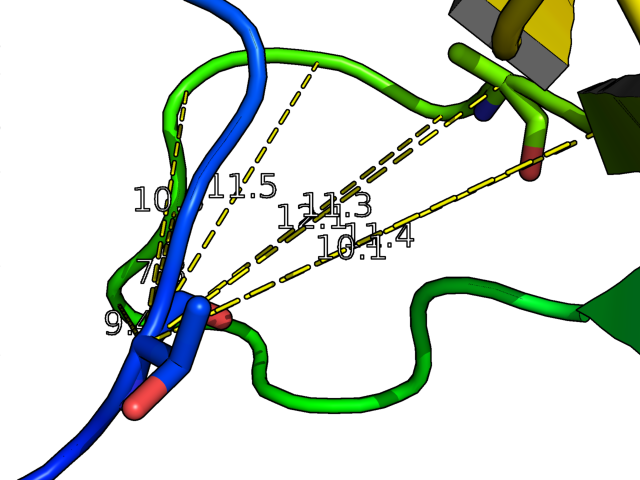
distance creates a new distance object between two selections. It will display all distances within the cutoff. Distance is also used to make hydrogen bonds.
Details
The distance wizard makes measuring distances easier than using the "dist" command for real-time operations. "dist" alone will show distances between selections (pk1) and (pk1), which can be set using the PkAt mouse action (usually CTRL-middle-click).
Usage
distance name = (selection1), (selection2) [,cutoff [,mode] ]
name
- string: name of the distance object to create
selection1
- string: first atom selection
selection2
- string: second atom selection
cutoff
- float: longest distance to show
mode
- 0: all interatomic distances
mode
- 1: only bond distances
mode
- 2: only show polar contact distances
PYMOL API
cmd.distance( string name, string selection1, string selection2,
string cutoff, string mode )
# returns the average distance between all atoms/frames
EXAMPLES
- Get and show the distance from residue 10's alpha carbon to residue 40's alpha carbon in 1ESR:
# make the first residue 0.
zero_residues 1esr, 0
distance i. 10 and n . CA, i. 40 and n. CA
- Get and show the distance from residue 10's alpha carbon to residue 35-42's alpha carbon in 1ESR:
# make the first residue 0.
zero_residues 1esr, 0
distance i. 10 and n . CA, i. 35-42 and n. CA
- This neat example shows how to create distance measurements from an atom in a molecule to all other atoms in the molecule (since PyMol supports wildcards).
cmd.dist("(/mol1///1/C)","(/mol1///2/C*)")
or written without the PyMolScript code,
dist /mol1///1/C, /mol1///2/C*
- Create multiple distance objects
for at1 in cmd.index("resi 10"): \
for at2 in cmd.index("resi 11"): \
cmd.dist(None, "%s`%d"%at1, "%s`%d"%at2)
distance (selection1), (selection2)
# example
dist i. 158 and n. CA, i. 160 and n. CA
distance mydist, 14/CA, 29/CA
distance hbonds, all, all, 3.2, mode=2