File list
Jump to navigation
Jump to search
This special page shows all uploaded files.
| Date | Name | Thumbnail | Size | User | Description | Versions |
|---|---|---|---|---|---|---|
| 16:06, 8 February 2011 | Map set ex.png (file) | 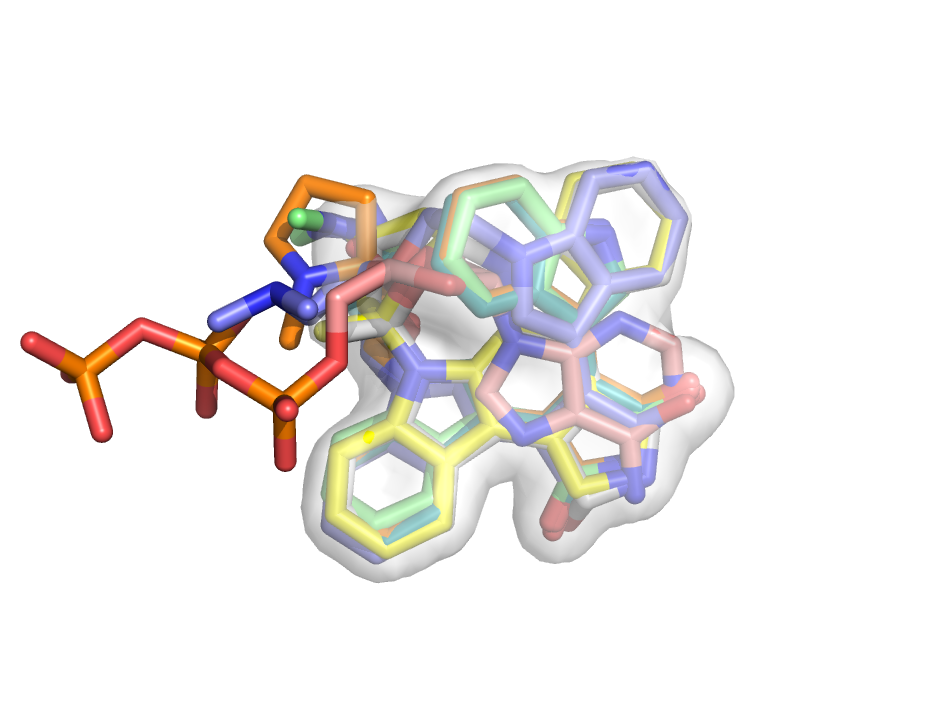 |
229 KB | Inchoate | Example consensus volume of the aligned ligands. | 1 |
| 10:39, 1 February 2011 | Apbs ex.png (file) | 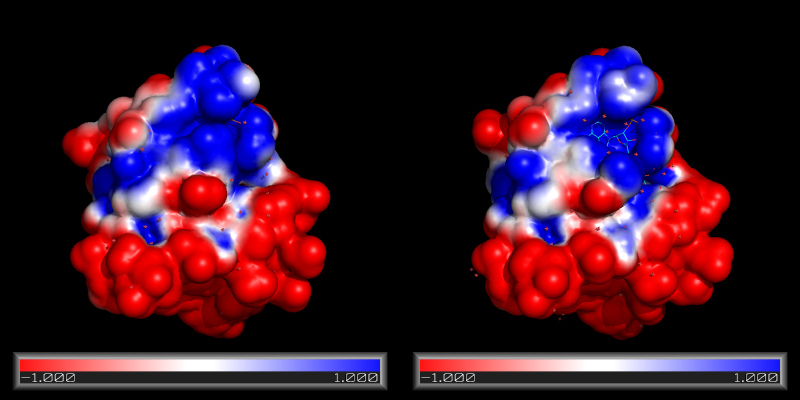 |
164 KB | Inchoate | Side-by-side map comparisons | 1 |
| 17:31, 24 January 2011 | AngewChemIntEd Comm Cover R Cighetti.jpg (file) | 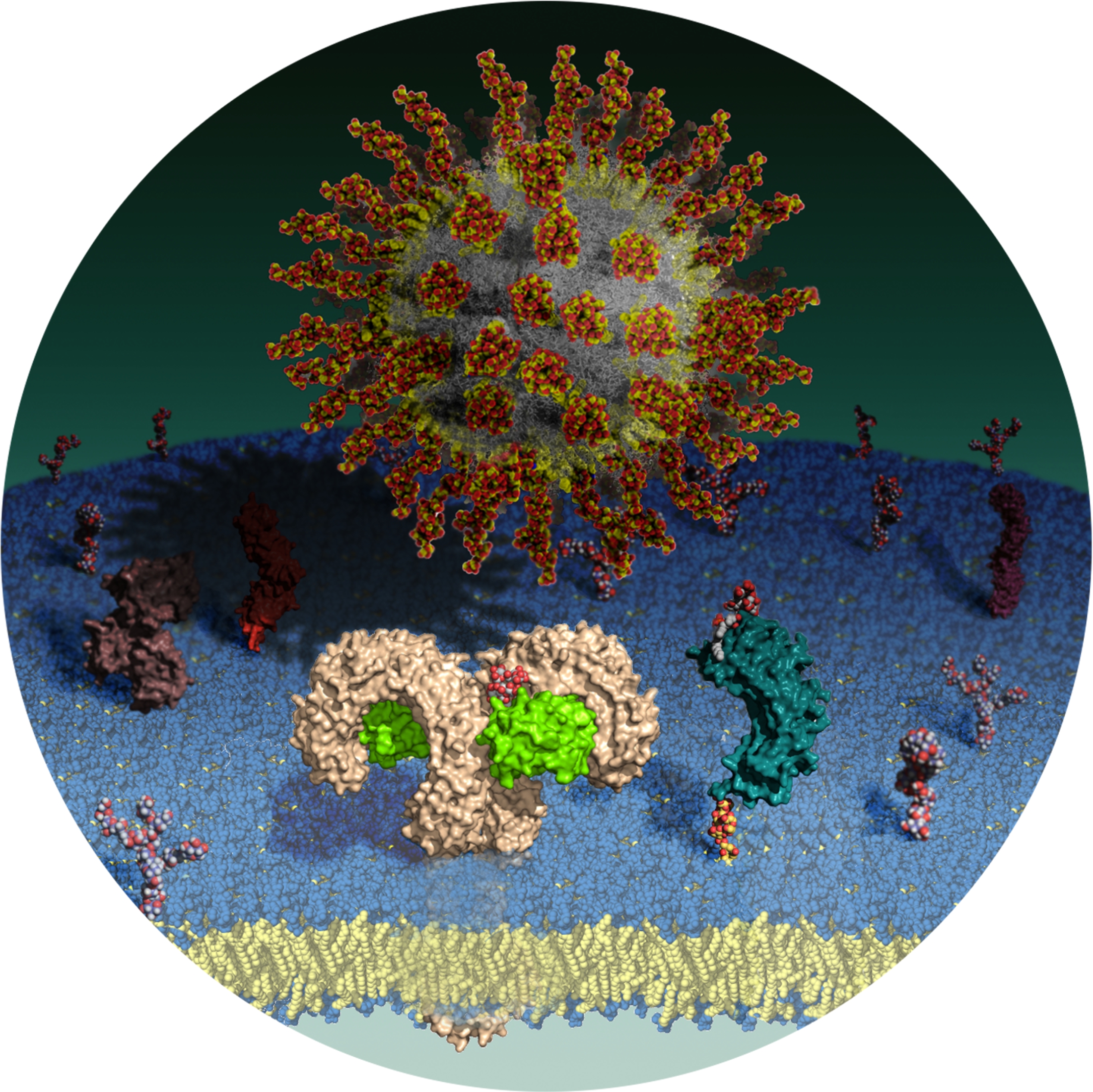 |
2.16 MB | Cigo86 | Image realized by Roberto Cighetti with PyMOL (Version 1.2r1) and GIMP (Version 2.6.8) http://onlinelibrary.wiley.com/doi/10.1002/anie.201004655/abstract | 1 |
| 17:03, 24 January 2011 | R Cighetti AngewChemCover.gif (file) | 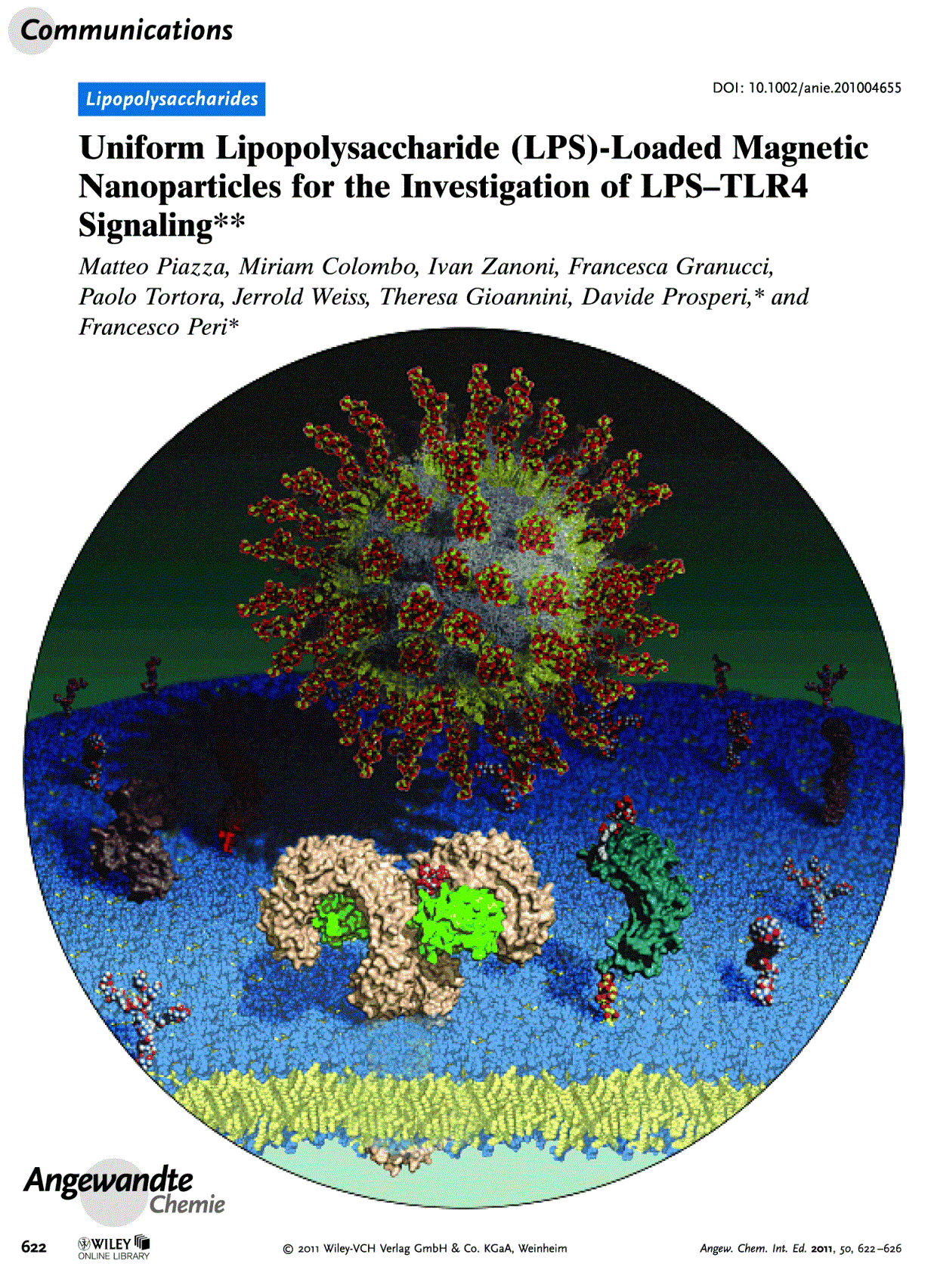 |
602 KB | Cigo86 | Communication Cover on Angewandte Chemie, 2011, 50, 622-626 Picture made by Roberto Cighetti with PyMOL (The PyMOL Molecular Graphics System, Version 1.2r1, Schrödinger, LLC) and GIMP (GNU Image Manipulation Program, version 2.6.8) from PDB ID: 3FXI (for | 1 |
| 12:15, 7 January 2011 | O5NT-C5NT-CA-dist-menu.png (file) | 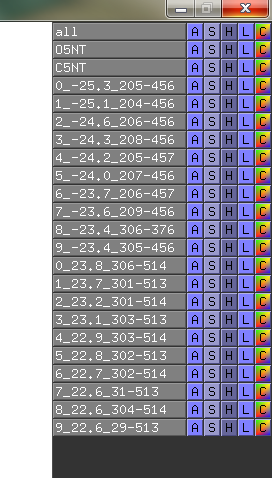 |
14 KB | Tlinnet | DisplacementMap parses best suggestions back to pymol as new objects. Command used: DisplacementMap O5NT, C5NT, CA, 40.0, 15.0, 10 | 1 |
| 12:11, 7 January 2011 | O5NT-C5NT-CA-dist.png (file) | 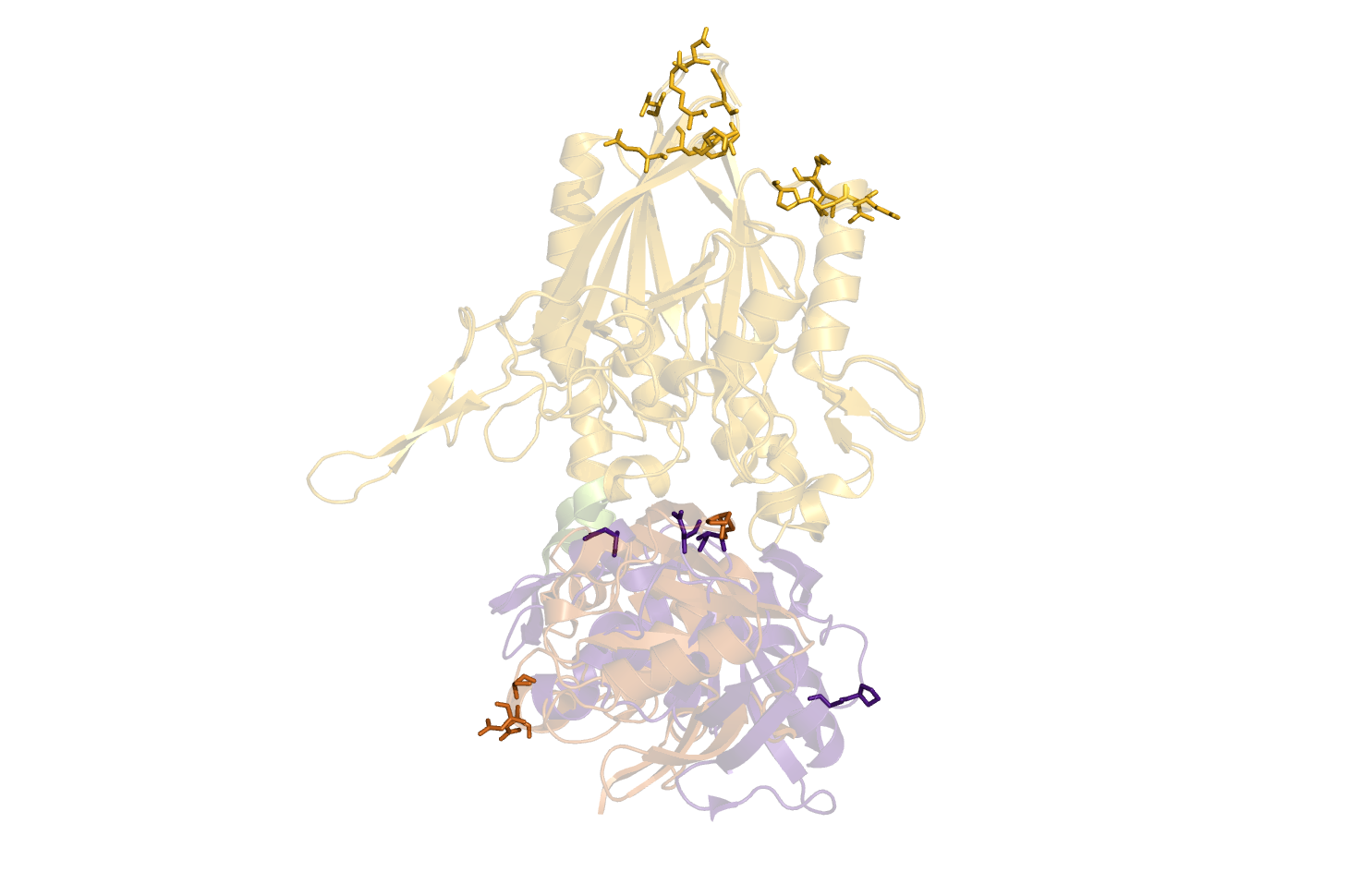 |
328 KB | Tlinnet | DisplacementMap parses best suggestions back to pymol as new objects. Command used: DisplacementMap O5NT, C5NT, CA, 40.0, 15.0, 10 | 1 |
| 12:07, 7 January 2011 | O5NT-C5NT-CA-dist-gnuplot.png (file) | 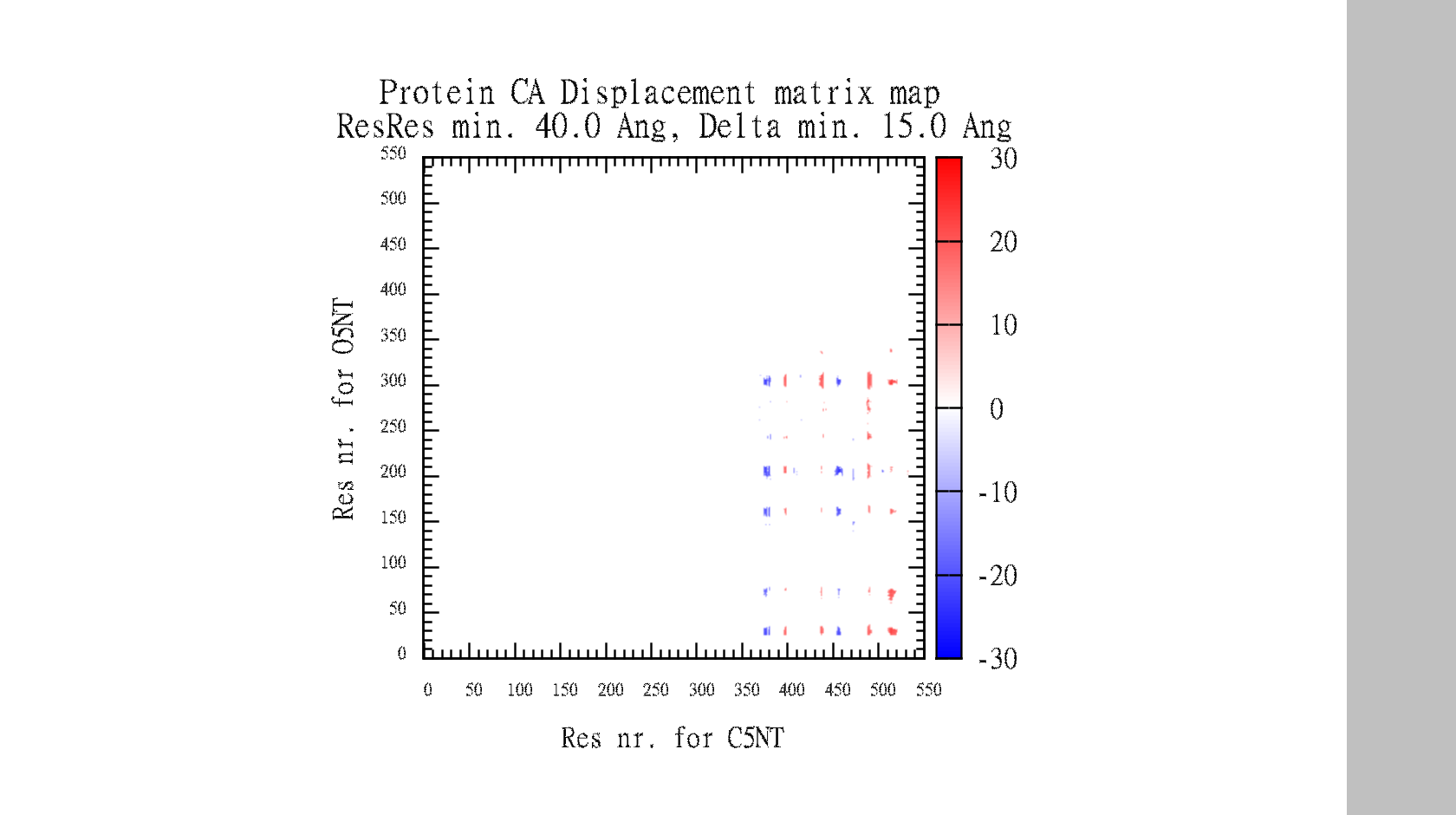 |
70 KB | Tlinnet | DisplacementMap gnuplot of data-matrix. Command: DisplacementMap O5NT, C5NT, CA, 40.0, 15.0, 10 | 1 |
| 17:44, 5 January 2011 | ColorByDisplacement-All-2.png (file) | 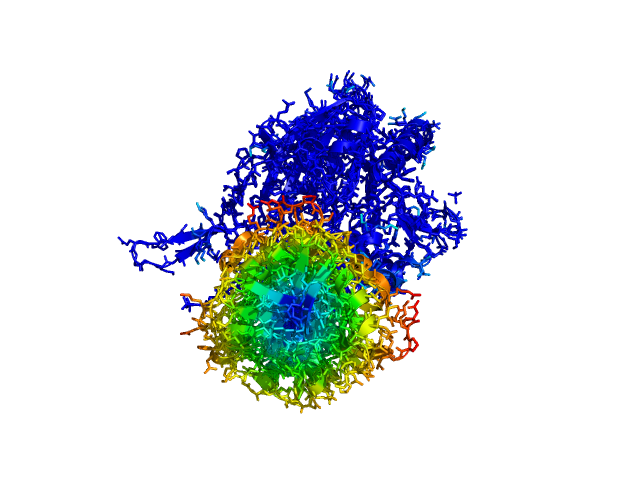 |
152 KB | Tlinnet | ColorByDisplacementAll used on 5'-nucleotidase (pdb:1HP1 and pdb:1HPU) | 1 |
| 17:43, 5 January 2011 | ColorByDisplacement-All-1.png (file) |  |
174 KB | Tlinnet | ColorByDisplacementAll used on 5'-nucleotidase (pdb:1HP1 and pdb:1HPU) | 1 |
| 17:29, 5 January 2011 | ColorByDisplacement-CA-2.png (file) |  |
98 KB | Tlinnet | ColorByDisplacementCA used on 5'-nucleotidase (pdb:1HP1 and pdb:1HPU) | 1 |
| 17:28, 5 January 2011 | ColorByDisplacement-CA-1.png (file) |  |
110 KB | Tlinnet | ColorByDisplacement used on 5'-nucleotidase (pdb:1HP1 and pdb:1HPU) | 1 |
| 16:30, 3 January 2011 | PLoS BiomolGraph 2010 InputFiles.tar.bz2 (file) | 2.03 MB | Cmura | 1 | ||
| 19:14, 26 December 2010 | O5NT-1HP1-A-C5NT-1HPU-C-CA-dist-map.png (file) | 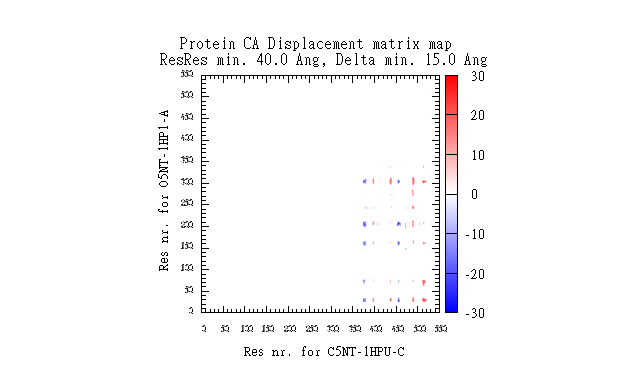 |
11 KB | Tlinnet | DisplacementMap gnuplot of data-matrix | 2 |
| 19:07, 26 December 2010 | O5NT-1HP1-A-C5NT-1HPU-C-CA-dist.png (file) |  |
106 KB | Tlinnet | DisplacementMap pymol output | 1 |
| 12:51, 2 December 2010 | Anaglyph1.png (file) |  |
346 KB | Inchoate | 1 | |
| 04:54, 9 November 2010 | Ukazky-plugin.png (file) |  |
12 KB | Hci | 1 | |
| 04:53, 9 November 2010 | Ukazky.png (file) |  |
621 KB | Hci | 1 | |
| 04:52, 9 November 2010 | Ukazky-plugin.PNG (file) |  |
12 KB | Hci | 1 | |
| 04:35, 9 November 2010 | Caver plugin2.png (file) |  |
18 KB | Hci | 1 | |
| 04:35, 9 November 2010 | Caver plugin1.png (file) |  |
30 KB | Hci | 1 | |
| 23:27, 19 October 2010 | 1rmv fiber.png (file) | 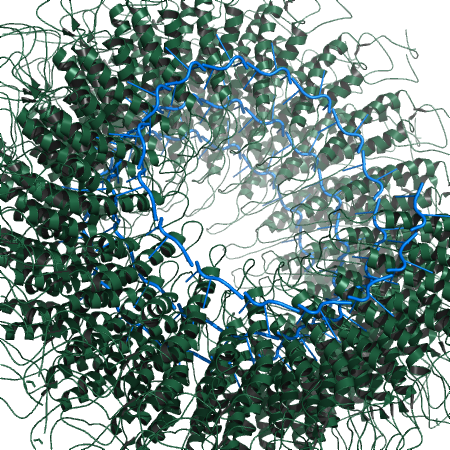 |
427 KB | Inchoate | 1RMV fiber, showing BiologicalUnit at work. | 1 |
| 05:00, 8 October 2010 | Orthoon2.png (file) |  |
66 KB | Karo | set orthoscopic, on (PDB code: 3AEH) | 1 |
| 04:59, 8 October 2010 | Orthooff2.png (file) |  |
66 KB | Karo | set orthoscopic, off (PDB code: 3AEH) | 1 |
| 04:58, 8 October 2010 | Orthoon.png (file) |  |
49 KB | Karo | set orthoscopic, on (PDB code 3AEH) | 1 |
| 04:56, 8 October 2010 | Orthooff.png (file) |  |
55 KB | Karo | 2 | |
| 21:40, 5 October 2010 | MMcoverMay2009.png (file) |  |
12 KB | MW | 1 | |
| 21:33, 5 October 2010 | MMcoverMay2009.gif (file) |  |
10 KB | MW | 1 | |
| 11:47, 29 September 2010 | PyMOLreferenceCard.zip (file) | 112 KB | Speleo3 | fix some commas fix 'near' and 'within' fix cartoon commands | 2 | |
| 11:32, 29 September 2010 | PyMOLreferenceCard-test.zip (file) | 68 KB | Inchoate | test | 1 | |
| 07:37, 29 September 2010 | PymolRef.pdf (file) | 112 KB | Speleo3 | fix some commas fix 'near' and 'within' fix cartoon commands | 2 | |
| 17:49, 28 September 2010 | Rtg10.png (file) |  |
94 KB | Inchoate | zoomed in and ray trace gain, 10 | 1 |
| 17:49, 28 September 2010 | Rtg0.png (file) |  |
74 KB | Inchoate | ray trace gain, 0 | 1 |
| 17:49, 28 September 2010 | Rtgd.png (file) |  |
78 KB | Inchoate | ray trace gain, default | 1 |
| 17:45, 28 September 2010 | Rsch1.png (file) |  |
60 KB | Inchoate | ribbon side chain helper on | 1 |
| 17:45, 28 September 2010 | Rsch0.png (file) |  |
74 KB | Inchoate | Ribbon side chain helper off | 1 |
| 18:19, 15 September 2010 | Surface residues ex.png (file) |  |
666 KB | Inchoate | 1 | |
| 09:20, 13 September 2010 | Helix orientation2.png (file) |  |
76 KB | Inchoate | helix angle. | 1 |
| 09:15, 13 September 2010 | Helix orientation1.png (file) |  |
67 KB | Inchoate | example helix orientation | 1 |
| 13:29, 7 September 2010 | 1hiw orient2.png (file) |  |
567 KB | Jaredsampson | Demonstration of orient command with 1hiw, two trimers shown. | 1 |
| 13:15, 7 September 2010 | 1hiw orient.png (file) |  |
484 KB | Jaredsampson | Demonstration of orient command with NCS symmetry of PDB 1hiw. | 1 |
| 12:02, 25 August 2010 | Extend3.png (file) |  |
32 KB | Baker1 | select (1kao and resi 64 and n. cz) extend 3 | 1 |
| 12:02, 25 August 2010 | Extend1.png (file) |  |
32 KB | Baker1 | select (1kao and resi 64 and n. cz) extend 1 | 1 |
| 11:59, 25 August 2010 | Extend0.png (file) |  |
32 KB | Baker1 | select (1kao and resi 64 and n. cz) | 1 |
| 11:04, 21 July 2010 | SupercellExample.png (file) |  |
311 KB | Speleo3 | fetch 2x19 supercell 1,1,2,2x19,withmates=1 disable 2x19 as ribbon, m* | 1 |
| 16:33, 19 July 2010 | KscA.png (file) | 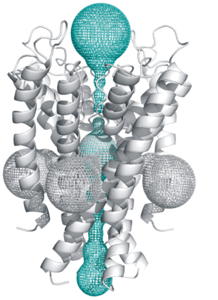 |
83 KB | Krapnik | MOLE found six channels in the open KscA K+ channel structure (PDB: 1k4c). From top one can distinguish the selectivity filter (with botlleneck radius 0.82 Å), cavity and channel gating (with bottleneck radius of 1.36 Å). Four perpendicular channels (le | 1 |
| 13:07, 30 June 2010 | BeCl2.png (file) | 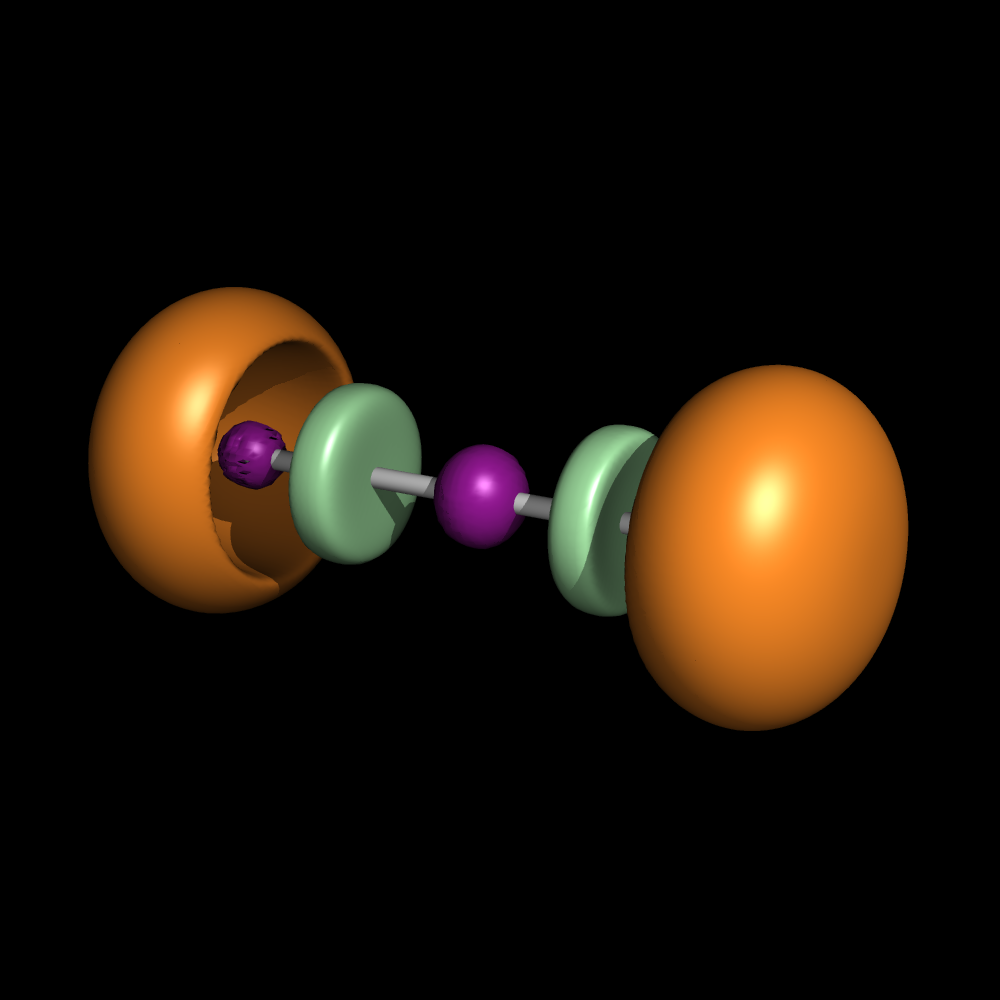 |
156 KB | Mretegan | Electron localization function for BeCl2 | 2 |
| 13:05, 30 June 2010 | H2O.png (file) | 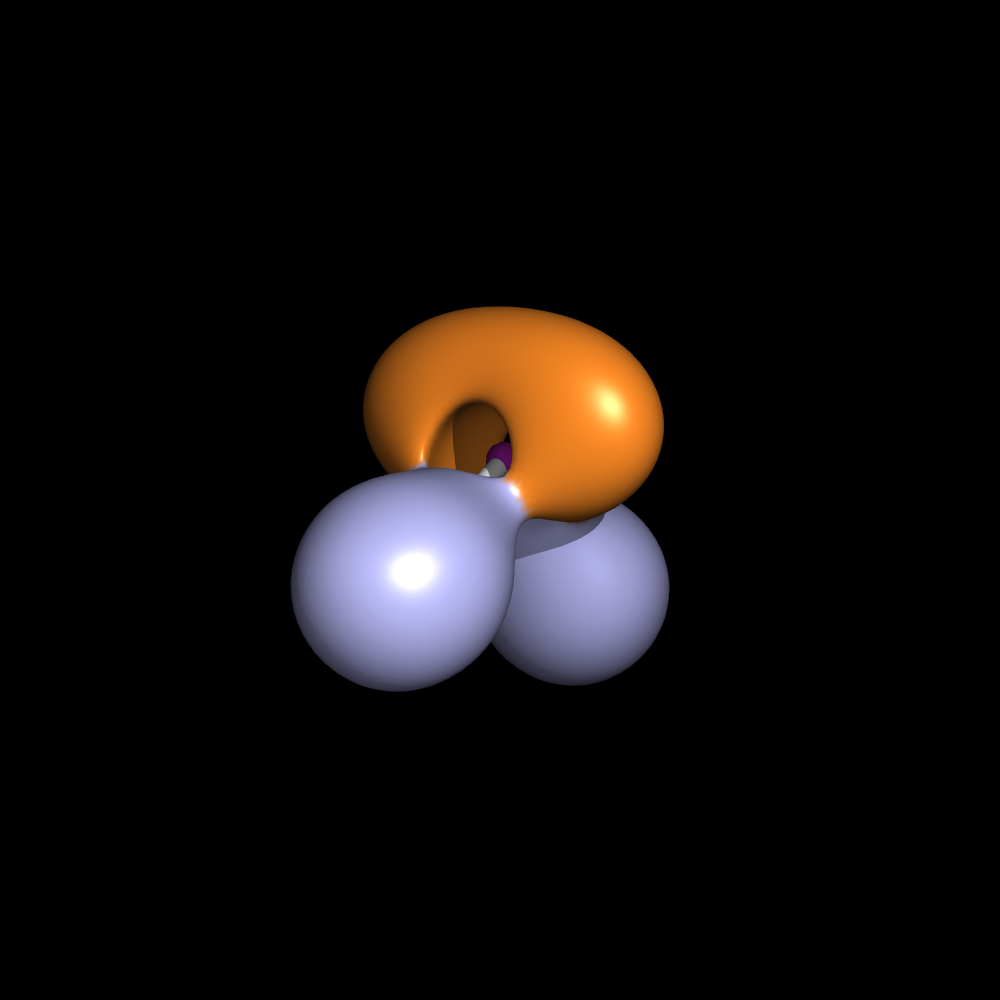 |
88 KB | Mretegan | Electron localization function for H2O | 1 |
| 18:13, 28 June 2010 | Surface by prop3.png (file) |  |
244 KB | Inchoate | 1 | |
| 18:13, 28 June 2010 | Surface by prop2.png (file) |  |
217 KB | Inchoate | 1 | |
| 18:13, 28 June 2010 | Surface by prop.png (file) |  |
546 KB | Inchoate | 1 |