Slice: Difference between revisions
Jump to navigation
Jump to search
mNo edit summary |
No edit summary |
||
| Line 3: | Line 3: | ||
== USAGE == | == USAGE == | ||
<source lang="python"> | <source lang="python"> | ||
slice_new name, map | slice_new name, map, [state, [source_state]] | ||
</source> | </source> | ||
| Line 9: | Line 9: | ||
<source lang="python"> | <source lang="python"> | ||
slice name, map | slice name, map, [state, [source_state]] | ||
</source> | </source> | ||
| Line 15: | Line 15: | ||
*'''name''' = the name for the new slice object (string) | *'''name''' = the name for the new slice object (string) | ||
*'''map''' = the name of the map object to use for computing the slice (string) | *'''map''' = the name of the map object to use for computing the slice (string) | ||
*'''state''' = the state into which the object should be loaded (default=1; set state=0 to append new mesh as a new state) | *'''state''' = the state into which the object should be loaded (default=1; set state=0 to append new mesh as a new state) | ||
*'''source_state''' = the state of the map from which the object should be loaded (default=0) | *'''source_state''' = the state of the map from which the object should be loaded (default=0) | ||
[[File:Slice_example_2_screenshot.png|200px|thumb|left|border|Screenshot of slice example #2]] | |||
<br clear="all" /> | |||
== EXAMPLES == | == EXAMPLES == | ||
<source lang="python"> | <source lang="python"> | ||
| Line 32: | Line 33: | ||
# Reset the view, to align view on XYZ axes | # Reset the view, to align view on XYZ axes | ||
reset | reset | ||
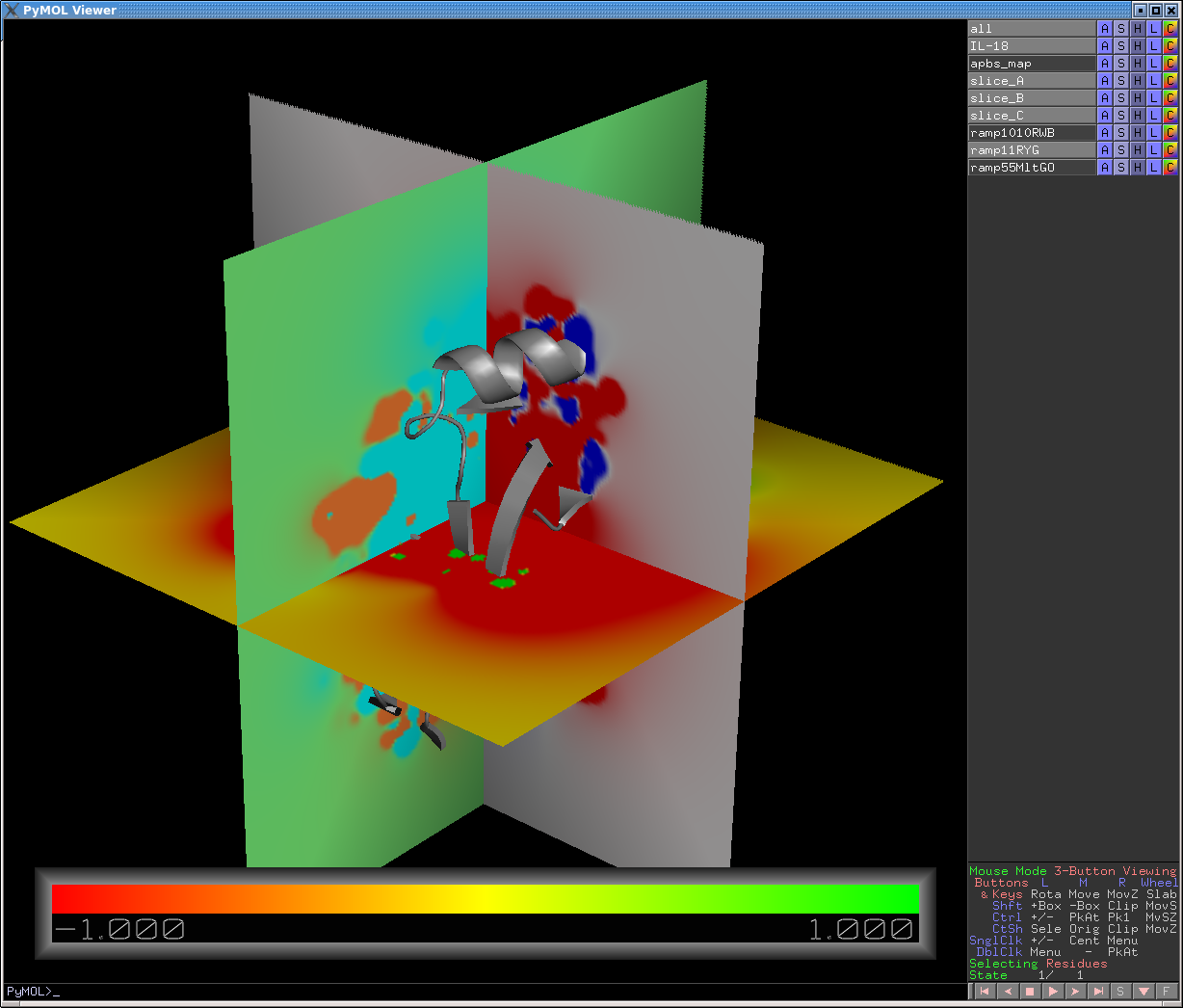
# Create a map slice perpendicular to current view. ( | # (Optional: Adjust view direction to your liking) | ||
# Create a map slice *perpendicular* to the current view. | |||
# The slice seems to be in the center of the APBS (or other) volmap. Map "tracking" is off by default. | |||
slice slice_A, apbs_map | slice slice_A, apbs_map | ||
# Rotate camera 90 degrees about the vertical axis | # Rotate camera 90 degrees about the vertical axis | ||
| Line 53: | Line 56: | ||
color ramp55MltGO, slice_C | color ramp55MltGO, slice_C | ||
# The result | # Adjust the fineness of the slice color gradations: | ||
cmd.set('slice_grid',0.1) # normally at 0.3; much finer than 0.05 gets a bit slow | |||
# Map slices can be moved, relative to other fixed objects (e.g., your protein/DNA/RNA), | |||
# by turning tracking on (Action menu), and the using the Shift-MouseWheel to move | |||
# the slice forward in and backward in Z. Adjust fineness of this Z-motion with: | |||
# cmd.set('mouse_wheel_scale',0.05) # normally at 0.5 | |||
# The result is shown in the image above. | |||
</source> | </source> | ||
== PYMOL API == | == PYMOL API == | ||
| Line 64: | Line 73: | ||
== NOTES == | == NOTES == | ||
Mis-identified as "slice_map" in documentation. [[slice]] or [[slice|slice_new]] is the correct command. Documentation also mentions two optional parameters which seem to be no longer supported (opacity = opacity of the new slice, and resolution = the number of pixels per sampling). | |||
== SEE ALSO == | == SEE ALSO == | ||
Revision as of 16:45, 16 March 2011
slice creates a slice object from a map object.
USAGE
slice_new name, map, [state, [source_state]]
or
slice name, map, [state, [source_state]]
ARGUMENTS
- name = the name for the new slice object (string)
- map = the name of the map object to use for computing the slice (string)
- state = the state into which the object should be loaded (default=1; set state=0 to append new mesh as a new state)
- source_state = the state of the map from which the object should be loaded (default=0)
EXAMPLES
# Create a map slice plane perpendicular to current view
slice a_new_slice, a_map
# A more complicated example that shows how to create multiple slices
# (in this case, 3 slices perpendicular to each other), each colored
# with a different color ramp and different contour levels:
# Reset the view, to align view on XYZ axes
reset
# (Optional: Adjust view direction to your liking)
# Create a map slice *perpendicular* to the current view.
# The slice seems to be in the center of the APBS (or other) volmap. Map "tracking" is off by default.
slice slice_A, apbs_map
# Rotate camera 90 degrees about the vertical axis
turn y, 90
# Second, perpendicular, slice
slice slice_B, apbs_map
# Rotate again, this time about the horizontal
turn x, 90
# Third slice
slice slice_C, apbs_map
# Define new color ramps: ramp_name, map_object, list of low/mid/hi values, 3 RGB triplets
ramp_new ramp1010RWB, apbs_map, [-10,0,10], [ [1,0,0], [1,1,1], [0,0,1] ]
ramp_new ramp11RYG, apbs_map, [-1,0,1], [ [1,0,0], [1,1,0], [0,1,0] ]
ramp_new ramp55MltGO, apbs_map, [-5,0,5], [ [0,1,1], [0.5,1,0.5], [1,0.5,0.2] ]
# Color the map slices
color ramp1010RWB, slice_A
color ramp11RYG, slice_B
color ramp55MltGO, slice_C
# Adjust the fineness of the slice color gradations:
cmd.set('slice_grid',0.1) # normally at 0.3; much finer than 0.05 gets a bit slow
# Map slices can be moved, relative to other fixed objects (e.g., your protein/DNA/RNA),
# by turning tracking on (Action menu), and the using the Shift-MouseWheel to move
# the slice forward in and backward in Z. Adjust fineness of this Z-motion with:
# cmd.set('mouse_wheel_scale',0.05) # normally at 0.5
# The result is shown in the image above.
PYMOL API
cmd.slice_new(string slice_name, string map_name, integer opacity=1, integer resolution=5, integer state=0, integer source_state=0)
NOTES
Mis-identified as "slice_map" in documentation. slice or slice_new is the correct command. Documentation also mentions two optional parameters which seem to be no longer supported (opacity = opacity of the new slice, and resolution = the number of pixels per sampling).
SEE ALSO
References
- PyMOL Source code