Main Page: Difference between revisions
Jump to navigation
Jump to search
m (Modified drawMinBoundingBox) |
No edit summary |
||
| Line 40: | Line 40: | ||
{| align="center" width="90%" style="background: #fafafa; border-right: 1px solid #333; border-left: 1px solid #333; border-bottom: 1px solid #333" | {| align="center" width="90%" style="background: #fafafa; border-right: 1px solid #333; border-left: 1px solid #333; border-bottom: 1px solid #333" | ||
|+ style="text-align: left; font-weight:bold; font-size:150%; color:#333; background: #EFE6FF; padding:10px; border: 1px solid #333" | News and Updates ([[Older_News|archive]]) | |+ style="text-align: left; font-weight:bold; font-size:150%; color:#333; background: #EFE6FF; padding:10px; border: 1px solid #333" | News and Updates ([[Older_News|archive]]) | ||
| | | | ||
{|style="background-color: transparent;" width="100%" | {|style="background-color: transparent;" width="100%" | ||
| Line 88: | Line 67: | ||
{|style="background-color: transparent;" width="100%" | {|style="background-color: transparent;" width="100%" | ||
! style="font-weight: bold; font-size: 110%; text-decoration: underline; color: #000; padding: 12px; text-align: left;" | Scripts & Plugins | ! style="font-weight: bold; font-size: 110%; text-decoration: underline; color: #000; padding: 12px; text-align: left;" | Scripts & Plugins | ||
|- | |||
| style="padding: 3px 5px 10px 15px;"| ♦ [[Pucker]] — finds the sugar pucker information (phase, amplitude, pucker) for a given selection. | |||
|- | |- | ||
| style="padding: 3px 5px 10px 15px;"| ♦ [[GetNamesInSel]] — find the names of all objects in a selection. | | style="padding: 3px 5px 10px 15px;"| ♦ [[GetNamesInSel]] — find the names of all objects in a selection. | ||
| Line 110: | Line 91: | ||
|} | |} | ||
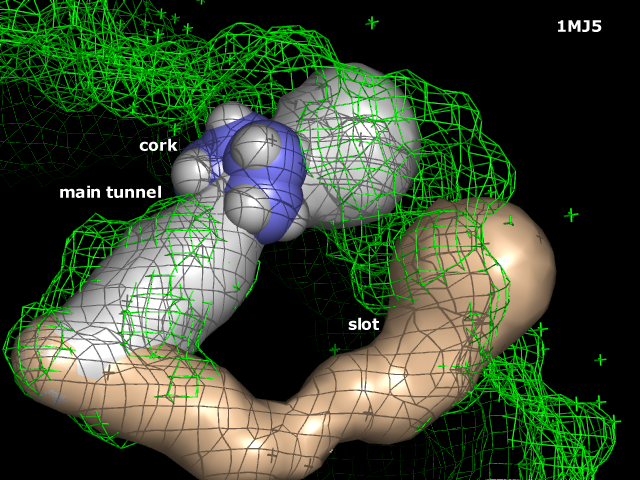
|width="150px" style="padding: 0 20px 20px 0; text-align:left" |[[Image:DrawMinBB.png|165px]] Screenshot of [[DrawBoundingBox]] in action. | |width="150px" style="padding: 0 20px 20px 0; text-align:left" |[[Image:DrawMinBB.png|165px]] Screenshot of [[DrawBoundingBox]] in action. | ||
|} | |||
{|style="background-color: transparent;" width="100%" | |||
|- | |||
|valign="top"| | |||
{|style="background-color: transparent;" width="100%" | |||
|- | |||
! style="font-weight: bold; font-size: 110%; text-decoration: underline; color: #000; padding: 12px; text-align: left;" | Wiki | |||
|- | |||
| style="padding: 3px 5px 10px 15px;"| ♦ Documented a few more settings. Also, check out the cool, [[Huge_surfaces]] page for handling very large objects and representing them as surfaces, in PyMOL. | |||
|- | |||
| style="padding: 3px 5px 10px 15px;"| ♦ We got our '''2,000,000th''' page view! | |||
|- | |||
| style="padding: 3px 5px 10px 15px;"| ♦ Added links in the table on the Main Page (above) for submitting & tracking bugs and feature reqeuests. | |||
|- | |||
| style="padding: 3px 5px 10px 15px;"| ♦ New logo for the wiki. It's DNA. You can easily see the major/minor grooves. If you don't see it, force a reload of the page (CTRL-F5, usually). | |||
|- | |||
| style="padding: 3px 5px 10px 15px;"| ♦ New category about PyMOL [[:Category:Performance|performance]]: making the impossible possible, and the difficult easier/faster. | |||
|} | |||
|valign="bottom" width="150px" style="padding: 0 20px 20px 0; clear:right;" |[[Image:080701_h.a.steinberg_biochemie.jpg|125px]] Sample Cover from the [[Covers]] gallery. | |||
|} | |} | ||
|} | |} | ||
Revision as of 17:38, 1 April 2009
| We are the community-based support site for the popular molecular visualization program, PyMOL. |
|
|||||||||||||||||||
| ||||||||||||||||||||||
|