MtsslDock: Difference between revisions
No edit summary |
|||
| Line 21: | Line 21: | ||
[[File:MtsslDock screenshot.jpg |640px]] | [[File:MtsslDock screenshot.jpg |640px]] | ||
==Installation== | |||
*Step 1: | *Step 1: | ||
Install the program by copying the code from the link above into an empty text file (e.g. "mtsslDock.py") located in the \Pymol\modules\pmg_tk\startup directory. After PyMOL has been started, the program can be launched from the WIZARDS menu. | Install the program by copying the code from the link above into an empty text file (e.g. "mtsslDock.py") located in the \Pymol\modules\pmg_tk\startup directory. After PyMOL has been started, the program can be launched from the WIZARDS menu. | ||
| Line 41: | Line 40: | ||
*[http://code.google.com/p/tkintertable/ tkintertable] | *[http://code.google.com/p/tkintertable/ tkintertable] | ||
==Docking== | |||
To do a docking run, label the two proteins with [[MtsslWizard | mtsslWizard (1.1)]] and then start mtsslDock from the Plugins menu. | To do a docking run, label the two proteins with [[MtsslWizard | mtsslWizard (1.1)]] and then start mtsslDock from the Plugins menu. | ||
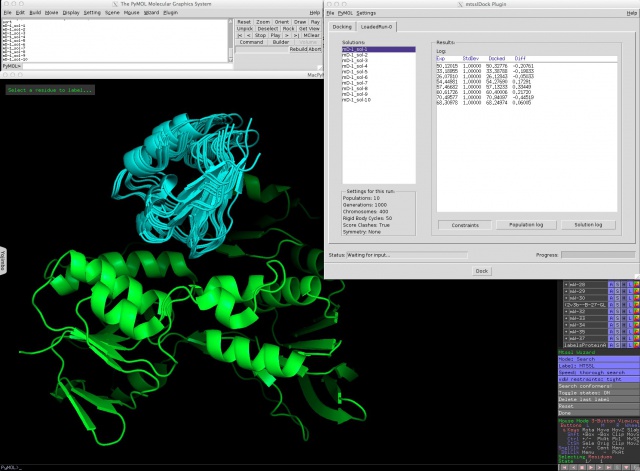
The GUI should then appear and look like this: | The GUI should then appear and look like this: | ||
| Line 63: | Line 62: | ||
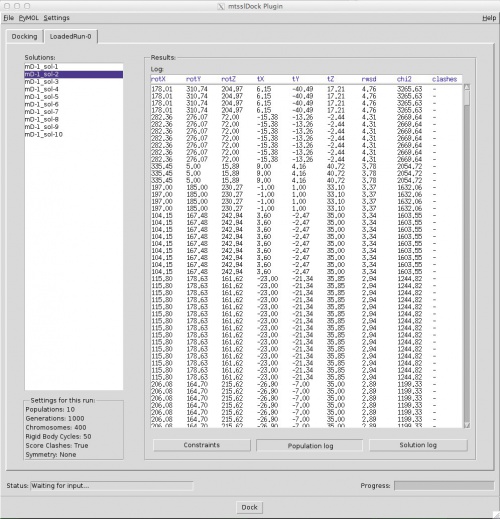
Here, the solutions are listed on the left, clashing ones are marked by "-clash" added to their names. The table on the right shows the log of the docking or the difference between docked and experimental distances for the selected solution, depending on what you select with the three buttons below the table. The box on the bottom left shows you the settings that were used for this docking run. | Here, the solutions are listed on the left, clashing ones are marked by "-clash" added to their names. The table on the right shows the log of the docking or the difference between docked and experimental distances for the selected solution, depending on what you select with the three buttons below the table. The box on the bottom left shows you the settings that were used for this docking run. | ||
==C2 symmetric structures, e.g. symmetric dimers== | ===C2 symmetric structures, e.g. symmetric dimers=== | ||
If you try the C2 dimer feature: Make a copy of your monomer and leave it at the exact same position. Then, label both monomers independently and import them as protein A and B. | If you try the C2 dimer feature: Make a copy of your monomer and leave it at the exact same position. Then, label both monomers independently and import them as protein A and B. | ||
Revision as of 11:20, 8 July 2013
| Type | PyMOL Plugin |
|---|---|
| Download | plugins/mtsslDockGui.py |
| Author(s) | Gregor Hagelueken |
| License | - |
| This code has been put under version control in the project Pymol-script-repo | |
mtsslDock
mtsslWizard is a PyMOL plugin for distance constrained rigid-body docking.
News
2013-07-04:
Version 1.0 in repository.
If you use this program, please reference:
Screen shot
Installation
- Step 1:
Install the program by copying the code from the link above into an empty text file (e.g. "mtsslDock.py") located in the \Pymol\modules\pmg_tk\startup directory. After PyMOL has been started, the program can be launched from the WIZARDS menu. Alternatively install the wizard via the plugins menu.
- Step 2:
This might be a bit tricky: In addition to scipy and numpy, you will need a module called tkintertable (http://code.google.com/p/tkintertable/). Download it and check the contents of the downloaded folder (It is probably called "tkintertable-1.1.2" or similar). There should be a folder in it that is called "tkintertable" put this folder into the 'site-packages' folder of the Python installation that PyMOL uses. On Windows it is usually the system-wide installation, on my Mac PyMOL uses its own Python: /Applications/PyMOLX11Hybrid.app/pymol/ext/lib/python2.7/site-packages
On my Mac I also had to install a new version of the Pmw module (for the GUI). You can get it here: http://sourceforge.net/projects/pmw/files/Pmw/Pmw.1.3.3/ Install it to /Applications/PyMOLX11Hybrid.app/pymol/modules into a folder called 'Pmw'. You might want to rename the original Pmw folder in case something gets messed up…
mtsslDock has been tested with PyMOL 1.5.
Additional requirements:
- SciPy
- NumPy
- tkintertable
Docking
To do a docking run, label the two proteins with mtsslWizard (1.1) and then start mtsslDock from the Plugins menu. The GUI should then appear and look like this:
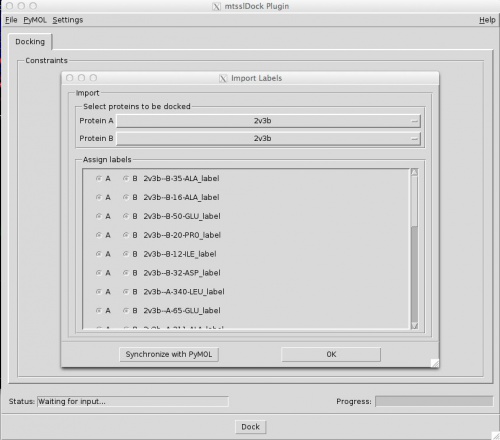
First select the two proteins that you want to dock using the pulldown menus at the top of the 'Import Labels' dialog. Protein A will be held fixed, protein B will be moved. In the list below these two pulldowns, the program will list all mtsslWizard labels that it found in the PyMOL session. You can assign each of them to either A or B using the radiobuttons. Once you are happy with everything, click "OK".
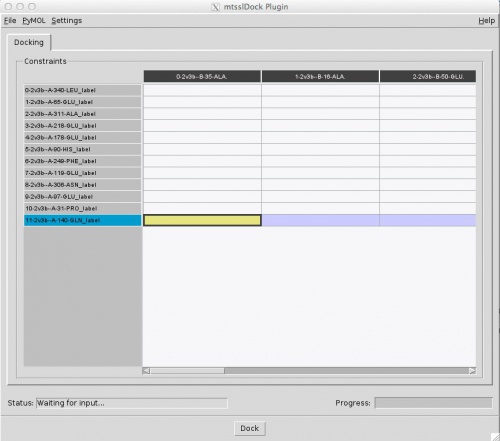
The program will then build up a table for the docking constraints and you can enter your distance data (the rows are proteinA labels, the columns protein B labels). If a distance was not measured, enter "0", otherwise enter the distance and uncertainty separated by semicolon, e.g. "12.4;5.0":
You can then save this table for later reference in the file menu. Note that if you do save it, you have to use the exact same order of labels if you want to use it again. If you want you can then change some docking settings in the 'Settings' menu, but the defaults should be fine.
Hit "Dock" at the bottom. The program will do 10 independent docking runs. Depending on the size of the molecules this might take 5-30 min. There is a progress bar on the bottom right. Once the docking has finished, a "Results" tab will be added to the GUI:
Here, the solutions are listed on the left, clashing ones are marked by "-clash" added to their names. The table on the right shows the log of the docking or the difference between docked and experimental distances for the selected solution, depending on what you select with the three buttons below the table. The box on the bottom left shows you the settings that were used for this docking run.
C2 symmetric structures, e.g. symmetric dimers
If you try the C2 dimer feature: Make a copy of your monomer and leave it at the exact same position. Then, label both monomers independently and import them as protein A and B.
The program was tested with a crystal structure of a protein:protein complex and it works well when you have at least 6 or better 8 distance constraints. See the paper for a discussion of performance.
Reference
If you find this program useful, please cite this paper:
It also contains detailed informations about the program, examples and a discussion of limitations of the approach.
Contact
hagelueken'at'pc.uni-bonn.de
Acknowledgement
Thanks to Jason Vertrees and Thomas Holder for some programming tips. Also to Damien Farrell for the tkintertable class.