Geo Measures Plugin: Difference between revisions
No edit summary |
No edit summary |
||
| Line 17: | Line 17: | ||
|} | |} | ||
== Video Tutorial == | |||
---- | |||
https://youtu.be/YKmIcU5weI0 | |||
---- | |||
== Installation == | == Installation == | ||
Latest revision as of 17:58, 23 December 2020
| Type | PyMOL Plugin |
|---|---|
| Download | https://github.com/lkagami/geo_measures_pymol/archive/master.zip |
| Author(s) | Luciano Porto Kagami, Gustavo Machado das Neves, Luís Fernando Saraiva Macedo Timmers, Rafael Andrade Caceres and Vera Lucia Eifler-Lima |
| License | GNU General Public License v3.0 |
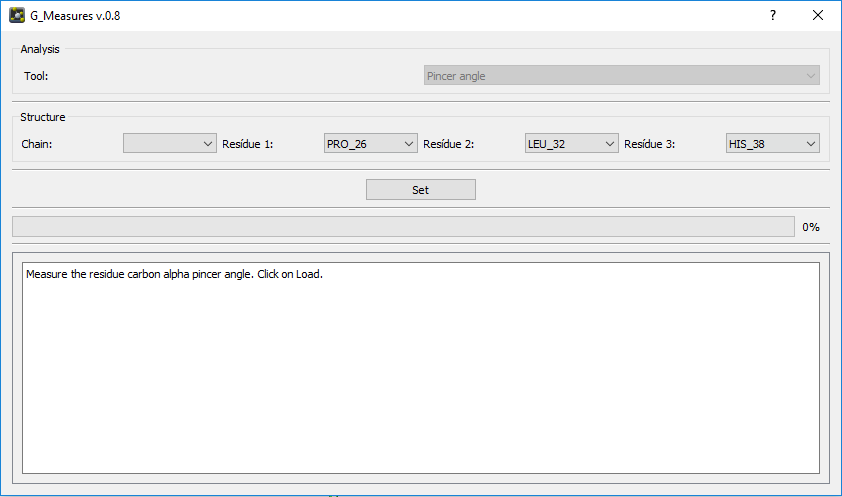
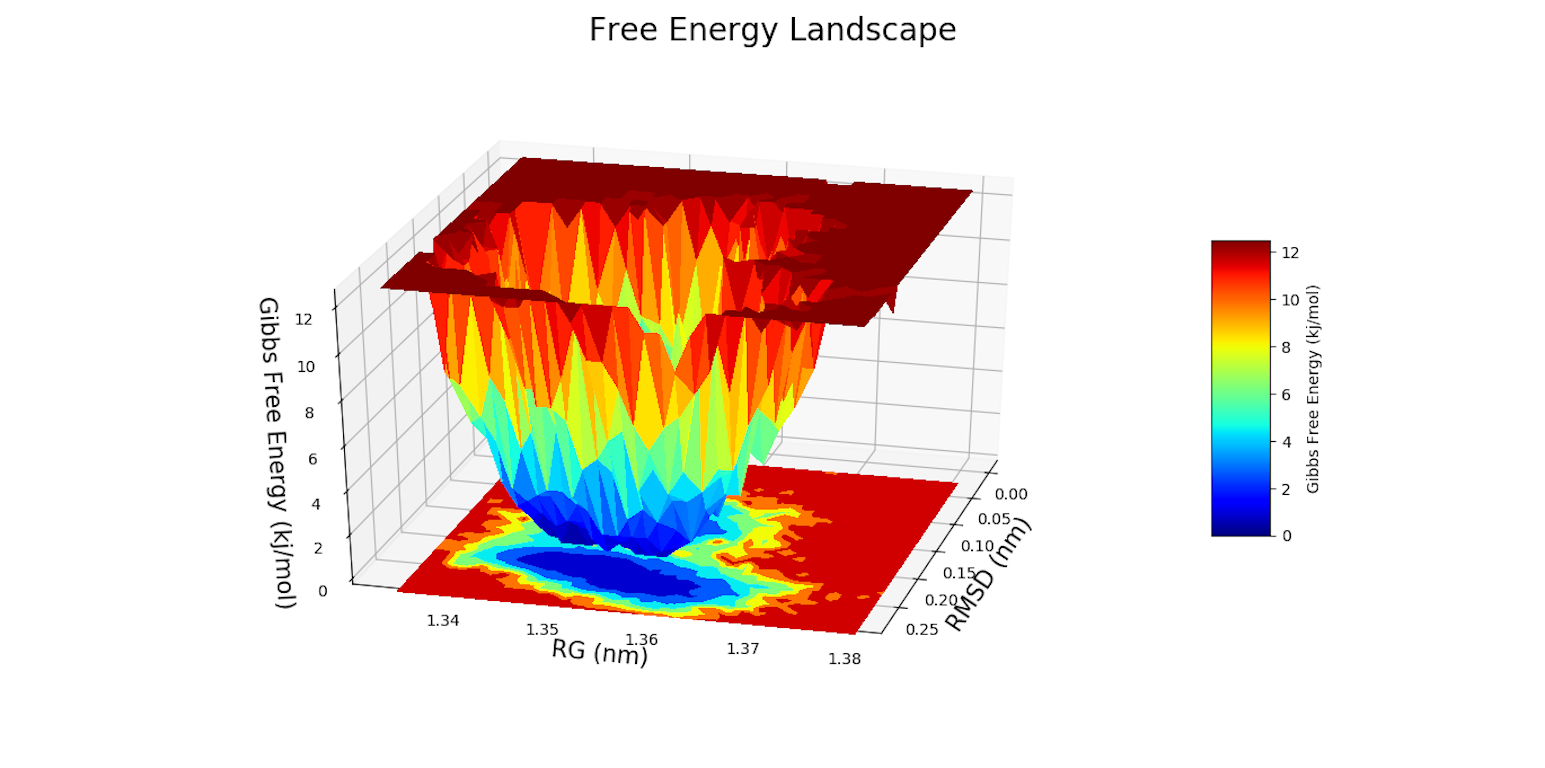
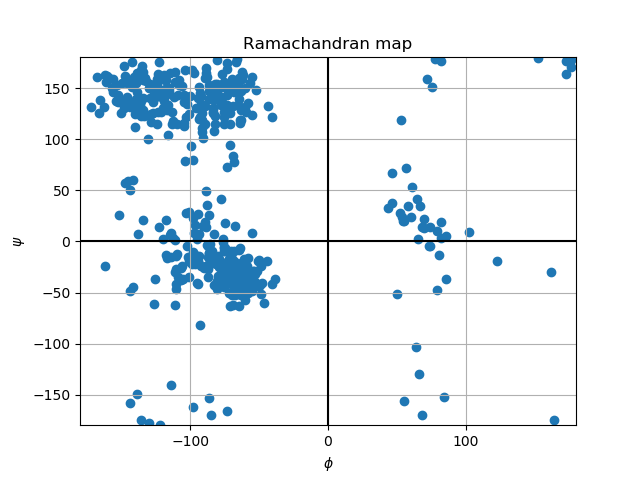
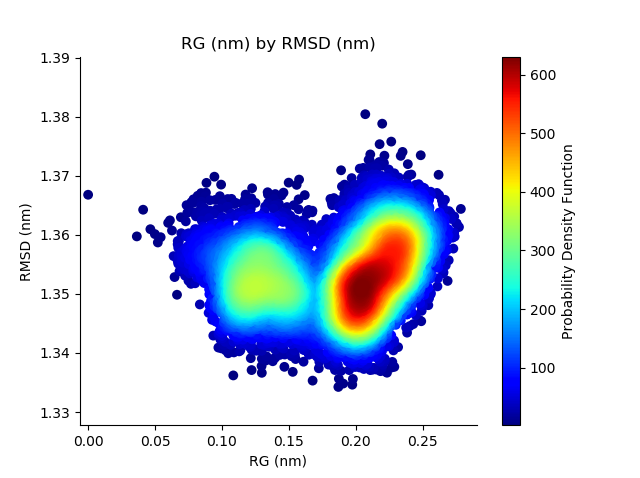
The Geo Measures Plugin can carry out geometric analysis on protein structures.In addition, it makes other trajectory analyzes such as Probability Density Function (PDF), Root Mean Square Deviation (RMSD), Radius of Gyration (RG), Free Energy Landscape (FEL), Principal Component Analysis (PCA), Ramachandran map, Root Mean Square Fluctuation (RMSF), Define Secondary Structure of Proteins (DSSP), and Modevectors. Altogether there are 14 tools, which can be easily used by the graphical interface.
Video Tutorial
Installation
Requirements:
PyMOL version higher than 2.0.
It is highly recommended to install PyMOL using the Ananconda3 package [1].
You need to install python packages.
To do this use the command:
pip install pandas matplotlib scipy mdtraj sklearn
Install geo_measures_pymol-master.zip using the PyMOL Plugin Manager
References
Kagami LP, das Neves GM, Timmers LFSM, Caceres RA,Eifler-Lima VL, Geo-Measures: A PyMOL plugin for protein structure ensembles analysis,Computational Biology and Chemistry(2020) doi.org/10.1016/j.compbiolchem.2020.107322
Universidade Federal do Rio Grande do Sul - Laboratório de Síntese Orgânica Medicinal - LaSOM