Caver2: Difference between revisions
No edit summary |
(Caver3) |
||
| (5 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
[ | <div style="background-color: #9f9; padding: 10px; margin-bottom: 20px; text-align: center"> | ||
[[Caver3|A new version of the Caver plugin is available]] | |||
</div> | |||
{{Infobox script-repo | |||
|type = plugin | |||
|filename = plugins/Caver2_1_2.py | |||
|author = [[User:Hci|CAVER 2.0]] | |||
|license = - | |||
}} | |||
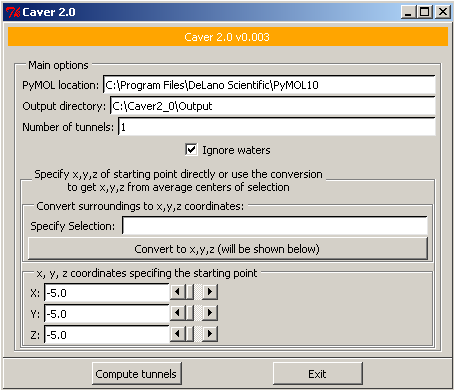
[http://loschmidt.chemi.muni.cz/caver/ CAVER 2.0] is a software tool for protein analysis leading to detection of channels. Channels are in fact void pathways leading from buried cavities to the outside solvent of a protein structure. Studying of these pathways is highly important in the area of drug design and molecular enzymology. | |||
[[File:ukazky-plugin.png]] | [[File:ukazky-plugin.png]] | ||
CAVER 2.0 provides rapid, accurate and fully automated calculation of channels not only for static molecules, but also for dynamic molecular simulations.<br> | |||
CAVER 2.0 provides rapid, accurate and fully automated calculation of channels not only for static molecules, but also for dynamic molecular simulations. | |||
CAVER 2.0 facilitates analysis of any molecular structure including proteins, nucleic acids or inorganic materials. | CAVER 2.0 facilitates analysis of any molecular structure including proteins, nucleic acids or inorganic materials. | ||
[[File:ukazky.png|1200px]] | [[File:ukazky.png|1200px]] | ||
CAVER 2.0 can be used in two possible ways. If you are accustomed with the PyMOL application, you can download the CAVER plugin for PyMOL. Then the standard functionality of PyMOL application for the subsequent exploration of computed channels can be used. The other possibility is to take advantage of our CAVER Viewer application, which has been designed exactly for the problem of detection and mainly for further exploration of channels. It includes the CAVER 2.0 algorithm for computation of channels in static molecule as well as in molecular dynamics. CAVER Viewer can be downloaded and installed as a standalone version or you can use the online version based on the Java Web Start technology. | CAVER 2.0 can be used in two possible ways. If you are accustomed with the PyMOL application, you can download the CAVER plugin for PyMOL. Then the standard functionality of PyMOL application for the subsequent exploration of computed channels can be used. The other possibility is to take advantage of our CAVER Viewer application, which has been designed exactly for the problem of detection and mainly for further exploration of channels. It includes the CAVER 2.0 algorithm for computation of channels in static molecule as well as in molecular dynamics. CAVER Viewer can be downloaded and installed as a standalone version or you can use the online version based on the Java Web Start technology. | ||
References: | References: | ||
| Line 24: | Line 26: | ||
Medek, Petr and Beneš, Petr and Sochor, Jiří,"[http://decibel.fi.muni.cz/download/papers/medek08.pdf Computation of tunnels in protein molecules using delaunay triangulation]". Journal of WSCG. Volume 15(1-3), 2007, Pages: 107-114. | Medek, Petr and Beneš, Petr and Sochor, Jiří,"[http://decibel.fi.muni.cz/download/papers/medek08.pdf Computation of tunnels in protein molecules using delaunay triangulation]". Journal of WSCG. Volume 15(1-3), 2007, Pages: 107-114. | ||
[[Category:Plugins]] | |||
[[Category:Pymol-script-repo]] | |||
Latest revision as of 02:20, 22 August 2017
| Type | PyMOL Plugin |
|---|---|
| Download | plugins/Caver2_1_2.py |
| Author(s) | CAVER 2.0 |
| License | - |
| This code has been put under version control in the project Pymol-script-repo | |
CAVER 2.0 is a software tool for protein analysis leading to detection of channels. Channels are in fact void pathways leading from buried cavities to the outside solvent of a protein structure. Studying of these pathways is highly important in the area of drug design and molecular enzymology.
CAVER 2.0 provides rapid, accurate and fully automated calculation of channels not only for static molecules, but also for dynamic molecular simulations.
CAVER 2.0 facilitates analysis of any molecular structure including proteins, nucleic acids or inorganic materials.
CAVER 2.0 can be used in two possible ways. If you are accustomed with the PyMOL application, you can download the CAVER plugin for PyMOL. Then the standard functionality of PyMOL application for the subsequent exploration of computed channels can be used. The other possibility is to take advantage of our CAVER Viewer application, which has been designed exactly for the problem of detection and mainly for further exploration of channels. It includes the CAVER 2.0 algorithm for computation of channels in static molecule as well as in molecular dynamics. CAVER Viewer can be downloaded and installed as a standalone version or you can use the online version based on the Java Web Start technology.
References:
Medek, Petr and Beneš, Petr and Kozlíková, Barbora and Chovancová, Eva and Pavelka, Antonín and Szabó, Tibor and Zamborský, Matúš and Andres, Filip and Klvaňa, Martin and Brezovský, Jan and Sochor, Jiří and Damborský, Jiří. "CAVER 2.0", 2008.
Medek, Petr and Beneš, Petr and Sochor, Jiří,"Computation of tunnels in protein molecules using delaunay triangulation". Journal of WSCG. Volume 15(1-3), 2007, Pages: 107-114.