APBS: Difference between revisions
Jump to navigation
Jump to search
(Added instructions for installing APBS from source) |
(update lots of outdated information) |
||
| (33 intermediate revisions by 13 users not shown) | |||
| Line 1: | Line 1: | ||
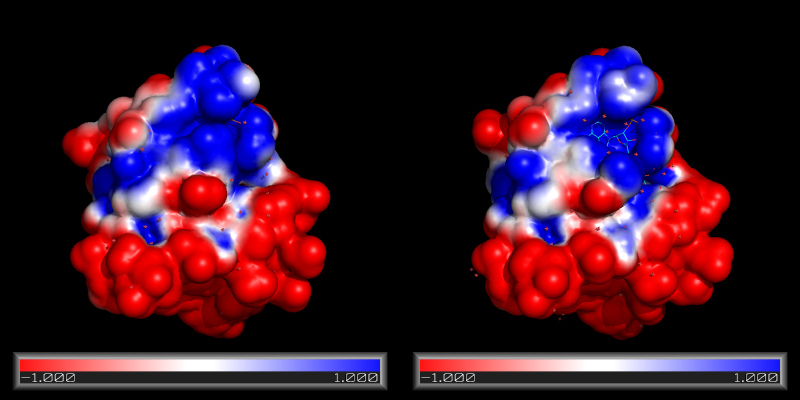
[[Image:Rna_surface_apbs.png|thumb|APBS-generated electrostatic surface displayed in PyMOL]] | [[Image:Rna_surface_apbs.png|thumb|APBS-generated electrostatic surface displayed in PyMOL]] | ||
[http:// | [http://www.poissonboltzmann.org/ APBS], the Adaptive Poisson-Boltzmann Solver, is a | ||
[http://www.oreilly.com/openbook/freedom/ freely] | |||
available macromolecular electrostatics calculation program released under a | |||
[http://apbs-pdb2pqr.readthedocs.io/en/latest/apbs/license.html BSD license]. | |||
PyMOL can display the results of the calculations as an electrostatic potential molecular surface. | |||
== APBS Plugins for PyMOL == | |||
* [[APBS Electrostatics Plugin]], included in [https://pymol.org/ Incentive PyMOL 2.0] | |||
* [[apbsplugin|APBS Tools 2.1]], based on the original version by [[User:Mglerner|Michael Lerner]]. | |||
Both plugins make it possible to run APBS from within PyMOL, and then display | |||
the results as a color-coded electrostatic surface (units <math>K_bT/e_c</math>) | |||
in the molecular display window (as with the image to the right). | |||
==Required Dependencies== | ==Required Dependencies== | ||
The plugins require '''apbs''' and '''pdb2pqr'''. | |||
* [https://pymol.org Incentive PyMOL] ships preconfigured with apbs and pdb2pqr | |||
* [https://github.com/Electrostatics/apbs-pdb2pqr/releases Official release downloads] for all platforms | |||
* Precompiled packages are also available in Ubuntu, Debian, Gentoo, [https://www.macports.org/ MacPorts], [http://www.finkproject.org/ Fink], and many other Unix-like distributions. Example for installation on Ubuntu: | |||
apt-get install apbs pdb2pqr | |||
After all components are installed, open the plugin and browse for apbs and pdb2pqr on the "Program Locations" tab (if they haven't been found automatically). | |||
== Troubleshooting == | |||
* ''(this might be outdated information):'' If the B-factor is <math>\geq 100,</math> then APBS doesn't properly read in the PDB file and thus outputs garbage (or dies). To fix this, set all b factors to be less than 100. <source lang="python">alter all, b=min(b,99.9)</source> The problem stems from how to parse a PDB file. The PDB file originally was written when most people used FORTRAN programs, and so the file format was specified by columns, not by the more modern comma separated value format we tend to prefer today. For the latest on the PDB format see the [http://www.wwpdb.org/docs.html new PDB format docs]. | |||
* APBS has problems, sometimes, in reading atoms with '''alternate conformations'''. You can remove the alternate locations with a simple script [[removeAlt]]. | |||
* For pdb2pqr, '''RNA''' resdiue names must be RA, RC, RG, and RU.<source lang="python">alter polymer & resn A+C+G+U, resn = "R" + resn</source> | |||
* Incomplete Residues: Some truncated PDB files include a single backbone atom of the next residue, e.g. [https://www.rcsb.org/structure/2xwu 2xwu] chain B residue 954 atom N. '''pdb2pqr''' reports: ''Error encountered: Too few atoms present to reconstruct or cap residue LEU B 954 in structure!''. The easiest solution is to remove that atom:<source lang="python">remove /2xwu//B/954</source> | |||
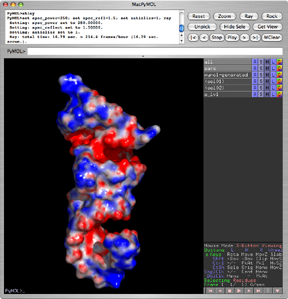
[[Image:Apbs_ex.png|thumb|right|300px|PyMOL visualizing two maps at once]] | |||
==Using APBS== | |||
See [[APBS Electrostatics Plugin]] and [[apbsplugin|APBS Tools2.1]]. | |||
[[Category:Electrostatics]] | |||
[[Category:Biochemical_Properties]] | |||
[[Category:Plugins]] | |||
Latest revision as of 14:05, 26 July 2018
APBS, the Adaptive Poisson-Boltzmann Solver, is a freely available macromolecular electrostatics calculation program released under a BSD license. PyMOL can display the results of the calculations as an electrostatic potential molecular surface.
APBS Plugins for PyMOL
- APBS Electrostatics Plugin, included in Incentive PyMOL 2.0
- APBS Tools 2.1, based on the original version by Michael Lerner.
Both plugins make it possible to run APBS from within PyMOL, and then display the results as a color-coded electrostatic surface (units ) in the molecular display window (as with the image to the right).
Required Dependencies
The plugins require apbs and pdb2pqr.
- Incentive PyMOL ships preconfigured with apbs and pdb2pqr
- Official release downloads for all platforms
- Precompiled packages are also available in Ubuntu, Debian, Gentoo, MacPorts, Fink, and many other Unix-like distributions. Example for installation on Ubuntu:
apt-get install apbs pdb2pqr
After all components are installed, open the plugin and browse for apbs and pdb2pqr on the "Program Locations" tab (if they haven't been found automatically).
Troubleshooting
- (this might be outdated information): If the B-factor is then APBS doesn't properly read in the PDB file and thus outputs garbage (or dies). To fix this, set all b factors to be less than 100. The problem stems from how to parse a PDB file. The PDB file originally was written when most people used FORTRAN programs, and so the file format was specified by columns, not by the more modern comma separated value format we tend to prefer today. For the latest on the PDB format see the new PDB format docs.
alter all, b=min(b,99.9)
- APBS has problems, sometimes, in reading atoms with alternate conformations. You can remove the alternate locations with a simple script removeAlt.
- For pdb2pqr, RNA resdiue names must be RA, RC, RG, and RU.
alter polymer & resn A+C+G+U, resn = "R" + resn
- Incomplete Residues: Some truncated PDB files include a single backbone atom of the next residue, e.g. 2xwu chain B residue 954 atom N. pdb2pqr reports: Error encountered: Too few atoms present to reconstruct or cap residue LEU B 954 in structure!. The easiest solution is to remove that atom:
remove /2xwu//B/954
Using APBS
See APBS Electrostatics Plugin and APBS Tools2.1.