MtsslWizard: Difference between revisions
No edit summary |
No edit summary |
||
| (71 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
{{Infobox script-repo | |||
|type = plugin | |||
|filename = plugins/mtsslWizard.py | |||
|author = [[User:Gha|Gregor Hagelueken]] | |||
|license = - | |||
}} | |||
= mtsslWizard = | = mtsslWizard = | ||
mtsslWizard is a PyMOL wizard for ''in silico'' spin labeling of proteins. | mtsslWizard is a PyMOL wizard for ''in silico'' spin labeling of proteins. | ||
==News== | |||
2020-05-21 | |||
*If you are having trouble with installation give [http://www.mtsslsuite.isb.ukbonn.de this] a try | |||
2017-01-17: | |||
*[http://www-hagelueken.thch.uni-bonn.de/index.php/en/2014-08-04-13-28-34/mtssldock Version 2.0 available]! | |||
2015-07-27: | |||
*uploaded a new version (1.3) to repository. Includes some new spin labels and a distance map feature. Plots can be viewed with [[mtsslPlotter]]. | |||
2014-03-04: | |||
* uploaded a new version (1.12) to repository. Fixes a bug that occurred on some machines. Thanks to C. Engelhard for noticing! | |||
2013-07-05: | |||
* added two new labels: DOTA and K1 (pAcPhe based) | |||
2013-01-31: | |||
Version 1.1 in script repository. | |||
Some new features of v1.1: | |||
* Much faster. The algorithm was optimized and the speed of the program is now almost independent off the size of the molecule. Due to this, 'thorough search' is now set as default and the 'copy and move' mode has been dropped. | |||
* New spin labels. The program now contains MTSSL, PROXYL and two labels for nucleic acids. Additional spin labels can be added upon request. | |||
* Distances and histogram are directly copied to the clipboard or written out to a file as before. The result file contains 4 columns: 1-distances, 2-bins for histogram, 3-histogram frequencies, 4-histogram frequencies with highest value normalized to 1.0 for comparisons | |||
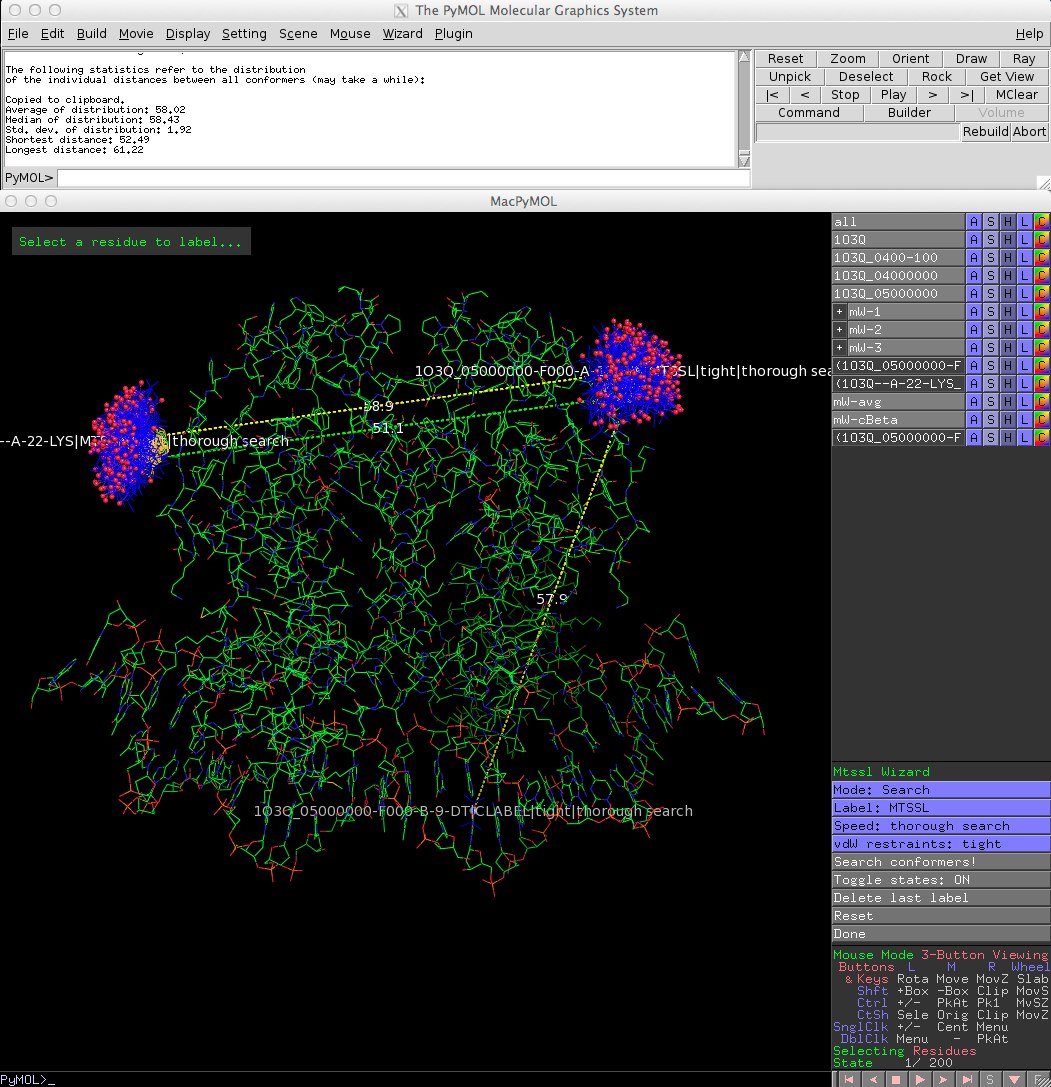
* Improved interface. Amongst other things, the clash control settings were simplified: There are now only two settings for vdW restraints: 'tight' and 'loose'. Also, only the average and c-Beta distances are displayed in the PyMOL viewer to avoid screen clutter (see screenshot). | |||
The new version was thoroughly tested. If you encounter any bugs or problems, please use the contact address below. | |||
Version 1.0 is still available [https://github.com/Pymol-Scripts/Pymol-script-repo/blob/5935bdad82a480a4e004c8c902ef69dc75389ac8/plugins/mtsslWizard.py here]. | |||
==Program features== | ==Program features== | ||
Spin labels can be attached to any position of a protein or nucleic acid just by pointing and clicking. The program then searches the conformational space of the label and determines which conformations of the label do not clash with the macromolecule. In "distance mode", distances between label ensembles can be determined and exported. In "distance map" mode, the program quickly estimates the inter-label distances between all possible spin pairs in a protein molecule. The result is saved as a color coded distance map. It is also possible to calculate difference distance maps between two conformations of a protein. | |||
Please reference: | |||
[http://www.springerlink.com/content/7148j5k2ppk475wu/ '''MtsslWizard: In Silico Spin-Labeling and Generation of Distance Distributions in PyMOL''', Gregor Hagelueken, Richard Ward, James H. Naismith and Olav Schiemann, DOI: 10.1007/s00723-012-0314-0] | |||
==Screen shot== | ==Screen shot== | ||
[[File: | [[File:MtsslWizardv1-1.jpg]] | ||
==Installation== | |||
Install the program by copying the code from the link above into an empty text file (e.g. "mtsslWizard.py") located in the \Pymol\modules\pmg_tk\startup directory. After PyMOL has been started, the program can be launched from the WIZARDS menu. | |||
Alternatively install the wizard via the plugins menu. | |||
mtsslWizard has been tested with PyMOL 1.7. It does not work with PyMOL 1.3. Here are manuals on how to install it on [[MAC_Install | Mac]] or [[Windows_Install | Windows]]. | |||
Additional requirements: | |||
*SciPy has to be installed | |||
*Pyperclip or xerox have to be installed for the clipboard support. | |||
====How people got it to run...==== | |||
*On a Mac it is easiest to install Pymol, Numpy, WxPython, Scipy and Matplotlib via fink. | |||
"I installed Pymol 1.5 via MacPorts. After putting pyperclip in the python 2.6 library folder, I then downloaded mtsslWizard and used the plugin installer to incorporate it. This was very simple and trouble-free." | |||
==Usage== | ==Usage== | ||
=== | ===Labeling=== | ||
#Open a | #Open a structure in PyMOL and remove any unwanted solvent molecules or ligands. | ||
#Start the mtsslWizard via Wizards>mtsslWizard | #Start the mtsslWizard via Wizards>mtsslWizard | ||
#If needed, vary the search parameters | #If needed, vary the search parameters: "Speed" and "vdW restraints". 'Speed' determines how many conformations of the label will be generated and checked for clashes. The 'vdW restraints' parameter determines how stringent conformations are checked for clashes with the macromolecular surface. | ||
# | #Click "Search conformers!" to start the calculation. | ||
=== | |||
===Distance calculation=== | |||
#change "Mode" to "Distance" | #change "Mode" to "Distance" | ||
#click the first and second ensemble | #click the first and second ensemble | ||
#The program will now calculate all possible distances between the conformations in the two ensembles. If both ensembles have many conformations, this can take | #The program will now calculate all possible distances between the conformations in the two ensembles. If both ensembles have many conformations, this can take a couple of seconds! | ||
=== | #The calculated distributions can be viewed with [[mtsslPlotter]]. | ||
MtsslWizard: In silico Spin-Labeling and Generation of Distance Distributions in PyMOL. | Distances and a histogram are directly copied to the clipboard or written out to a file. The result file contains 4 columns: | ||
===Contact | #distances | ||
#bins for histogram | |||
== | #histogram frequencies | ||
#histogram frequencies with highest value normalized to 1.0 for comparisons | |||
===Distance maps=== | |||
#change "Mode" to "Distance map" | |||
#For a simple distance map: click twice on the protein of interest | |||
#For a difference distance map between two conformations of a protein: click once on each of the two conformations. For this feature to work properly, both conformations should have the same number of residues. This can be easily achieved using standard PyMOL commands. | |||
#The maps can be viewed with [[mtsslPlotter]] | |||
==Reference== | |||
If you find this program useful, please cite this paper: | |||
Hagelueken G, Ward R, Naismith JH, Schiemann O (2012) MtsslWizard: In silico Spin-Labeling and Generation of Distance Distributions in PyMOL. Appl. Magn. Res. accepted for publication | |||
It also contains detailed informations about the program, examples and a discussion of limitations of the approach. | |||
==Older versions== | |||
*[https://github.com/Pymol-Scripts/Pymol-script-repo/blob/5935bdad82a480a4e004c8c902ef69dc75389ac8/plugins/mtsslWizard.py Version 1.0] | |||
==Contact== | |||
hagelueken'at' uni-bonn.de | |||
==Acknowledgement== | |||
Thanks to Jason Vertrees and Thomas Holder for some programming tips. | |||
Latest revision as of 14:00, 21 May 2020
| Type | PyMOL Plugin |
|---|---|
| Download | plugins/mtsslWizard.py |
| Author(s) | Gregor Hagelueken |
| License | - |
| This code has been put under version control in the project Pymol-script-repo | |
mtsslWizard
mtsslWizard is a PyMOL wizard for in silico spin labeling of proteins.
News
2020-05-21
- If you are having trouble with installation give this a try
2017-01-17:
2015-07-27:
- uploaded a new version (1.3) to repository. Includes some new spin labels and a distance map feature. Plots can be viewed with mtsslPlotter.
2014-03-04:
- uploaded a new version (1.12) to repository. Fixes a bug that occurred on some machines. Thanks to C. Engelhard for noticing!
2013-07-05:
- added two new labels: DOTA and K1 (pAcPhe based)
2013-01-31:
Version 1.1 in script repository.
Some new features of v1.1:
- Much faster. The algorithm was optimized and the speed of the program is now almost independent off the size of the molecule. Due to this, 'thorough search' is now set as default and the 'copy and move' mode has been dropped.
- New spin labels. The program now contains MTSSL, PROXYL and two labels for nucleic acids. Additional spin labels can be added upon request.
- Distances and histogram are directly copied to the clipboard or written out to a file as before. The result file contains 4 columns: 1-distances, 2-bins for histogram, 3-histogram frequencies, 4-histogram frequencies with highest value normalized to 1.0 for comparisons
- Improved interface. Amongst other things, the clash control settings were simplified: There are now only two settings for vdW restraints: 'tight' and 'loose'. Also, only the average and c-Beta distances are displayed in the PyMOL viewer to avoid screen clutter (see screenshot).
The new version was thoroughly tested. If you encounter any bugs or problems, please use the contact address below.
Version 1.0 is still available here.
Program features
Spin labels can be attached to any position of a protein or nucleic acid just by pointing and clicking. The program then searches the conformational space of the label and determines which conformations of the label do not clash with the macromolecule. In "distance mode", distances between label ensembles can be determined and exported. In "distance map" mode, the program quickly estimates the inter-label distances between all possible spin pairs in a protein molecule. The result is saved as a color coded distance map. It is also possible to calculate difference distance maps between two conformations of a protein.
Please reference:
Screen shot
Installation
Install the program by copying the code from the link above into an empty text file (e.g. "mtsslWizard.py") located in the \Pymol\modules\pmg_tk\startup directory. After PyMOL has been started, the program can be launched from the WIZARDS menu. Alternatively install the wizard via the plugins menu.
mtsslWizard has been tested with PyMOL 1.7. It does not work with PyMOL 1.3. Here are manuals on how to install it on Mac or Windows.
Additional requirements:
- SciPy has to be installed
- Pyperclip or xerox have to be installed for the clipboard support.
How people got it to run...
- On a Mac it is easiest to install Pymol, Numpy, WxPython, Scipy and Matplotlib via fink.
"I installed Pymol 1.5 via MacPorts. After putting pyperclip in the python 2.6 library folder, I then downloaded mtsslWizard and used the plugin installer to incorporate it. This was very simple and trouble-free."
Usage
Labeling
- Open a structure in PyMOL and remove any unwanted solvent molecules or ligands.
- Start the mtsslWizard via Wizards>mtsslWizard
- If needed, vary the search parameters: "Speed" and "vdW restraints". 'Speed' determines how many conformations of the label will be generated and checked for clashes. The 'vdW restraints' parameter determines how stringent conformations are checked for clashes with the macromolecular surface.
- Click "Search conformers!" to start the calculation.
Distance calculation
- change "Mode" to "Distance"
- click the first and second ensemble
- The program will now calculate all possible distances between the conformations in the two ensembles. If both ensembles have many conformations, this can take a couple of seconds!
- The calculated distributions can be viewed with mtsslPlotter.
Distances and a histogram are directly copied to the clipboard or written out to a file. The result file contains 4 columns:
- distances
- bins for histogram
- histogram frequencies
- histogram frequencies with highest value normalized to 1.0 for comparisons
Distance maps
- change "Mode" to "Distance map"
- For a simple distance map: click twice on the protein of interest
- For a difference distance map between two conformations of a protein: click once on each of the two conformations. For this feature to work properly, both conformations should have the same number of residues. This can be easily achieved using standard PyMOL commands.
- The maps can be viewed with mtsslPlotter
Reference
If you find this program useful, please cite this paper:
Hagelueken G, Ward R, Naismith JH, Schiemann O (2012) MtsslWizard: In silico Spin-Labeling and Generation of Distance Distributions in PyMOL. Appl. Magn. Res. accepted for publication
It also contains detailed informations about the program, examples and a discussion of limitations of the approach.
Older versions
Contact
hagelueken'at' uni-bonn.de
Acknowledgement
Thanks to Jason Vertrees and Thomas Holder for some programming tips.