Caver: Difference between revisions
No edit summary |
(caver3) |
||
| (4 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
[[ | #REDIRECT [[caver3]] | ||
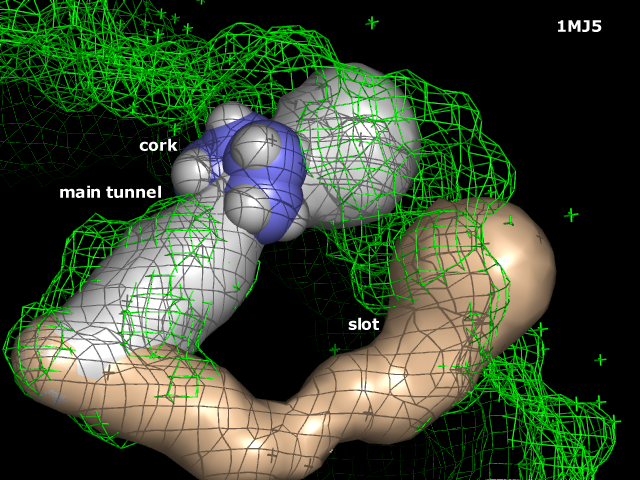
CAVER 1.0 provides rapid, accurate and fully automated calculation of pathways leading from buried cavities to outside solvent in static and dynamic protein structures. Study of these pathways is important in drug design and molecular enzymology to understand binding of inhibitors to the receptors, substrate binding and product egress from the enzyme active sites. Calculated pathways can be visualized by graphic program PyMol dissecting anatomy and dynamics of entrance tunnels. CAVER allows analysis of any molecular structure including proteins, nucleic acids, inorganic materials, etc. | A new version, CAVER 2.0, was recently developed. For further information see [http://www.pymolwiki.org/index.php/CAVER_2.0 CAVER 2.0 PyMOLWiki page] | ||
Previous CAVER 1.0 (no longer maintained) provides rapid, accurate and fully automated calculation of pathways leading from buried cavities to outside solvent in static and dynamic protein structures. Study of these pathways is important in drug design and molecular enzymology to understand binding of inhibitors to the receptors, substrate binding and product egress from the enzyme active sites. Calculated pathways can be visualized by graphic program PyMol dissecting anatomy and dynamics of entrance tunnels. CAVER allows analysis of any molecular structure including proteins, nucleic acids, inorganic materials, etc. | |||
References: | References: | ||
Latest revision as of 16:48, 27 July 2017
Redirect to:
A new version, CAVER 2.0, was recently developed. For further information see CAVER 2.0 PyMOLWiki page
Previous CAVER 1.0 (no longer maintained) provides rapid, accurate and fully automated calculation of pathways leading from buried cavities to outside solvent in static and dynamic protein structures. Study of these pathways is important in drug design and molecular enzymology to understand binding of inhibitors to the receptors, substrate binding and product egress from the enzyme active sites. Calculated pathways can be visualized by graphic program PyMol dissecting anatomy and dynamics of entrance tunnels. CAVER allows analysis of any molecular structure including proteins, nucleic acids, inorganic materials, etc.
References:
Petřek M., Otyepka M., Banáš P., Košinová P., Koča J. and Damborský J., CAVER: A New Tool to Explore Routes from Protein Clefts, Pockets and Cavities, BMC Bioinformatics 2006, 7: 316
Damborský J., Petřek M., Banáš P., Otyepka M., Identification of Tunnels in Proteins, Nucleic Acids, Inorganic Materials and Molecular Ensembles, Biotechnology Journal 2007, 2: 62-67