CavitOmiX
Appearance
| Type | PyMOL Plugin |
|---|---|
| Download | https://innophore.com/cavitomix |
| Author(s) | Georg Steinkellner, Christian C. Gruber, Karl Gruber*, and the Innophore Team |
| License | |
| https://innophore.com | |

CavitOmiX 1.0
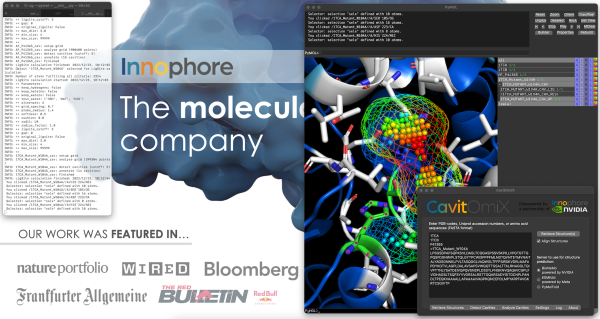
CavitOmiX plugin for Schrodinger’s PyMOL, a nifty tool that allows you to analyze protein cavities from any input structure. You can now dive deep into your proteins, cavities, and binding sites using crystal structures and state-of-the-art AI models from OpenFold (powered by NVIDIA’s BioNeMo service), DeepMind`s AlphaFold and ESMFold by Meta. Even more exciting: just enter any protein sequence and you will get the structure predicted by OpenFold or ESMFold loaded into your PyMOL within seconds.
- Catalophore™ Cavities can be calculated for molecules loaded in PyMOL
- Predict protein structures within seconds for any protein sequence using OpenFold by NVIDIA BioNeMo (coming soon!) and ESMFold by Meta
- AlphaFold models can be retrieved via UniProt ID
- Analyze the hydrophobicity of your Catalophore™ cavities
- Protein structures can be retrieved from the PDB using the PDB code
- Mix and match all the above in a single entry, align the structures and get a quick overview
- Catalophore™ cavity calculation settings can be changed
- Each Catalophore™ cavity is an "residue" entry and each cavity point is an "atom", so you can select, remove, copy, represent cavities to your liking!
Overview