Nonstandard Amino Acids
Jump to navigation
Jump to search
The printable version is no longer supported and may have rendering errors. Please update your browser bookmarks and please use the default browser print function instead.
Overview
This page talks a little about how PyMOL deals with nonstandard amino acids, and the various representations and options available.
By default, PyMOL considers nonstandard amino acids as HETERO atoms. Therefore, when you draw a default surface, the heteroatoms are not included. Also, nonstandard atoms are left out of the backbone representation in the cartoon and ribbon representations.
Workarounds
- If you're looking to represent the backbone via ribbon or cartoon, then just use the mutagenesis to modify the nonstandard amino acid to something standard, like GLU, or LEU. The draw/ray your structure as needed.
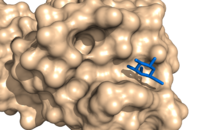
- If you want the nonstandard amino acid to be included in the surface representation, then set surface_mode to 1. For example, consider the images below. We have a hetero atom shown in the image at left, not included in the surface, become part of the surface when we set the surface_mode to 1.
- Surface Mode Examples
surface_mode set to 0, the default. The galactose (blue) is not considered part of the surface.
surface_mode set to 1 -- now including heteroatoms. The galactose and all heteroatoms (blue) are now considered part of the surface and colored blue.
- Another work around is to try
flag ignore, bymol polymer, set
rebuild