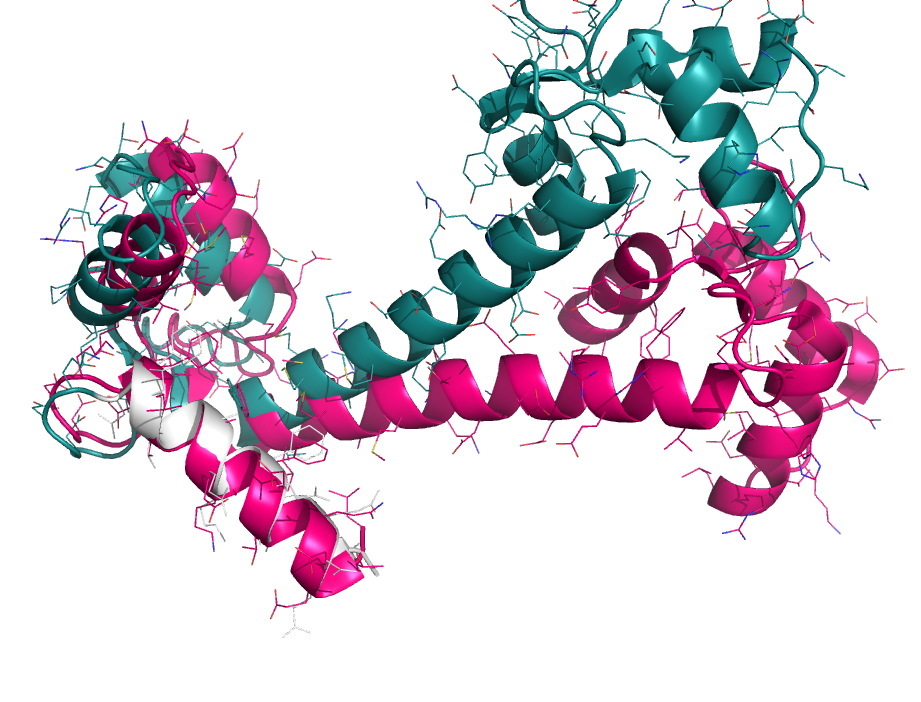
Focus alignment
Jump to navigation
Jump to search
Overview
When aligning binding sites, it's useful to focus on a particular subset of atoms. However, alignments can go haywire when choosing non-contiguous subsets of atoms. To get around this we first perform a sequence alignment to establish an initial pairing. Then, the user-specified sub-selection is used to pare down the equivalenced atoms for pair-fitting. See the examples.
Examples
focus_alignment 1cll, 1ggz, 1cll and i. 4-20
focus_alignment 2erk, 2b9f, 2erk and i. 50-75
The Code
# focus_alignment
#
# Author: Jason Vertrees
# Date : 08-17-2011
#
import pymol
# example usage
# focus_alignment 2erk, 2b9f, i. 50-70 and 2erk
# focus_alignment 1cll, 1ggz, 1cll and i. 4-20
def focus_alignment(obj1, obj2, sel, debug=False):
"""
PARAMS
obj1
structure 1
obj2
structure 2
sel
the selection from either obj1 or
obj2 to focus the pair_fitting on.
When providing this selection, please
make sure you also specify selected
atoms from ONE object.
NOTES
This function will first align obj1 and obj2 using
a sequence alignment. This creates a mapping of
residues from obj1 to obj2. Next, the selection, sel,
is used to find only those atoms in the alignment and
in sel. These atoms are paired with their mapped
atoms from the alignment in the other object. These
two subsets of atoms are then pair_fit to give
an optimal sub-alignment.
"""
aln = "aln"
_sel = "__sel"
ssel_model = ""
a1, a2, a_target, modelA, modelB, sel_model = [],[],[],[],[],[]
obj1, obj2 = "poly and " + obj1, "poly and " + obj2
# space dictionary for iterate
space = { 'a1' : a1,
'a2' : a2,
'a_target' : a_target,
'sel_model' : sel_model,
'modelA' : modelA,
'modelB' : modelB }
# initial unfocused alignment
cmd.align(obj1, obj2, cycles=0, object=aln)
# record the initial indices
s = "n. CA and (%s and %s)"
cmd.iterate(s % (obj1,aln), "a1.append(index)",space=space)
cmd.iterate(s % (obj2,aln), "a2.append(index)",space=space)
if debug:
print "# [debug] num atom in aln1 = ", len(a1)
print "# [debug] num atom in aln2 = ", len(a2)
# determine who owns the focused selection and
# get canonical object names
cmd.iterate("first %s" % sel, "sel_model.append(model)",space=space)
cmd.iterate("first %s" % obj1, "modelA.append(model)",space=space)
cmd.iterate("first %s" % obj2, "modelB.append(model)",space=space)
ssel_model = sel_model[0]
if debug:
print "# [debug] selection is in object %s" % ssel_model
# focus the target selection
cmd.iterate(sel + " and n. CA", "a_target.append(index)",space=space)
if debug:
print "# [debug] a_target has %d members." % len(a_target)
# select the correct object from which to index
target_list = None
if ssel_model==modelA[0]:
target_list = a1
elif sel_model==modelB[0]:
target_list = a2
else:
print "# error: selection on which to focus was not found"
print "# error: in either object passed in."
print sel_model
print modelA
print modelB
return False
id1, id2 = [], []
for x in a_target:
try:
idx = target_list.index(x)
if debug:
print "Current index: %d" % idx
id1.append( str(a1[idx]) )
id2.append( str(a2[idx]) )
except:
pass
if debug:
print "# [debug] id1 = %s" % id1
print "# [debug] id2 = %s" % id2
sel1 = "+".join(id1)
sel2 = "+".join(id2)
if debug:
print "# [debug] sel1 = %s" % sel1
cmd.pair_fit(obj1 + " and aln and index " + sel1,
obj2 + " and aln and index " + sel2)
cmd.extend("focus_alignment", focus_alignment)
print " A new function 'focus_alignment' was added to PyMOL."
print " Type 'help focus_alignment' if you need help."