Nonstandard Amino Acids
Jump to navigation
Jump to search
Overview
This page talks a little about how PyMOL deals with nonstandard amino acids, and the various representations and options available.
By default, PyMOL considers nonstandard amino acids as HETERO atoms. Therefore, when you draw a default surface, the heteroatoms are not included. Also, nonstandard atoms are left out of the backbone representation in the cartoon and ribbon representations.
Workarounds
- If you're looking to represent the backbone via ribbon or cartoon, then just use the mutagenesis to modify the nonstandard amino acid to something standard, like GLU, or LEU. The draw/ray your structure as needed.
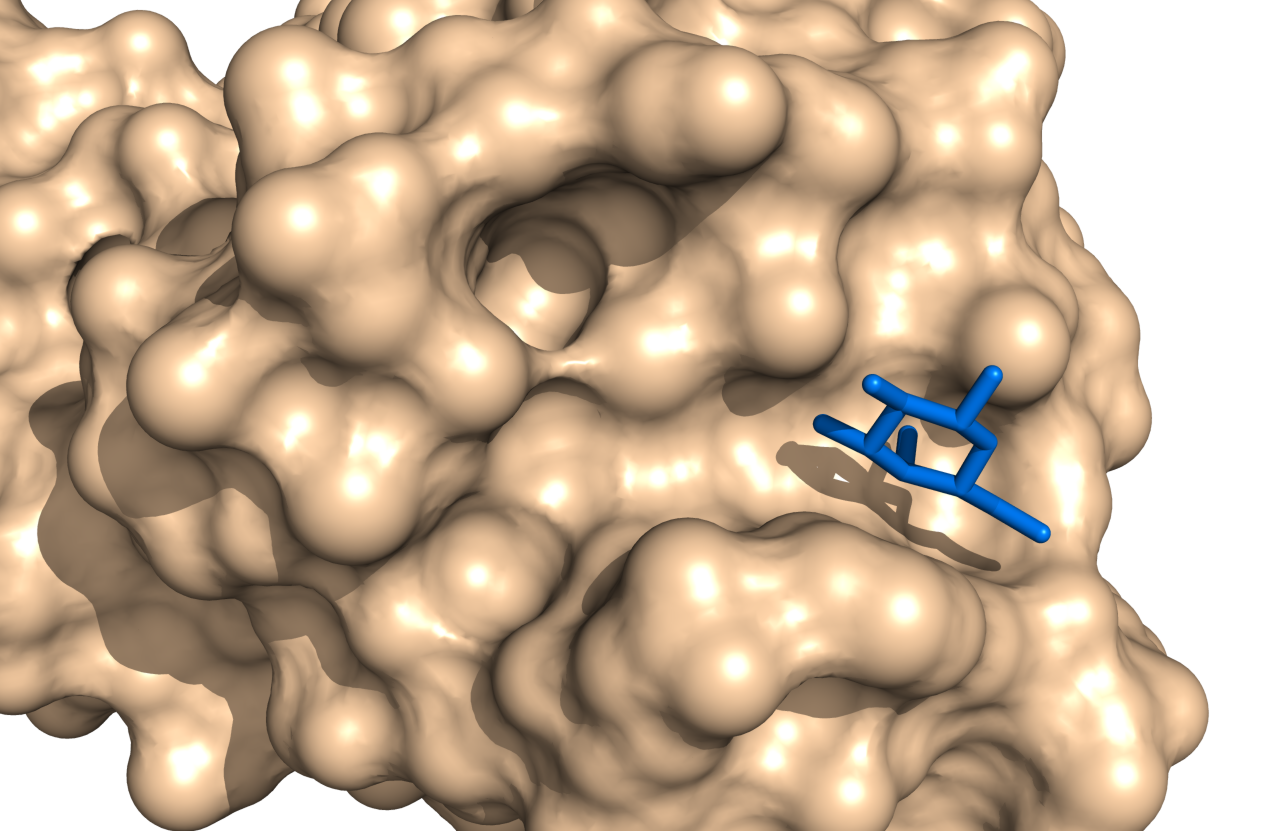
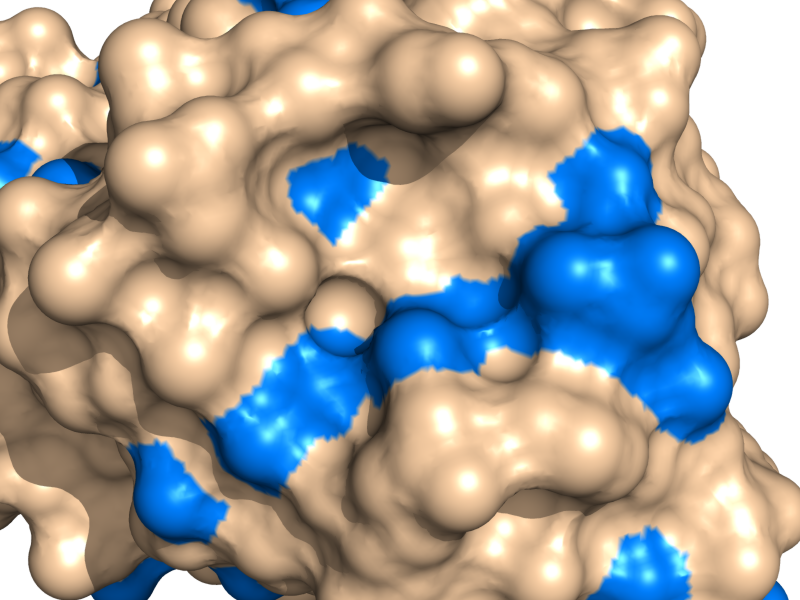
- If you want the nonstandard amino acid to be included in the surface representation, then set surface_mode to 1. For example, consider the images below. We have a hetero atom shown in the image at left, not included in the surface, become part of the surface when we set the surface_mode to 1.
- Surface Mode Examples
surface_mode set to 0, the default. The galactose (blue) is not considered part of the surface.
surface_mode set to 1 -- now including heteroatoms. The galactose and all heteroatoms (blue) are now considered part of the surface and colored blue.
- Another work around is to try
flag ignore, bymol polymer, set
rebuild
but, that didn't work for me.