PPIIMoL
PPIIMoL
PPIIMoL is a Python module for PyMOL that automates the detection of polyproline II (PPII) helices in proteins. It identifies PPII-like φ/ψ angle patterns, screens for plausible non-canonical Cα–H···O=C contacts, and provides one-click visualization and export.
This tool was developed as part of a Bachelor's Thesis in Computer Engineering in collaboration with the Protein Structure, Dynamics and Interactions by NMR Group at the Instituto de Química-Física “Blas Cabrera” (IQF-CSIC). The module’s design and validation take as a primary reference the data and architectonic principles reported by Segura Rodríguez & Laurents (2024) (see How to cite).
Scientific background
Polyproline II (PPII) helices are extended, left-handed motifs (≈3 residues/turn) typically enriched in glycine- and proline-rich domains. Although common in several glycine-rich bundles, they are often unannotated in PDB files. PPIIMoL automates their detection directly in PyMOL to improve speed and reproducibility.
Features
- 🔍 Automatic detection of PPII segments via phi/psi angle analysis.
- 🧬 Identification of Cα-H···O=C interactions relevant to structural stability.
- 📊 CSV export of detected segments and interactions.
- 🎨 Direct visualization in PyMOL with customizable color codes.
- 🖱️ Simple Tkinter-based GUI — no commands required; all actions are accessible via buttons.
Requirements
- PyMOL 2.x or newer.
- Python with Tkinter enabled (for the GUI).
Installation
Option A — Single-file download (simplest)
- Download `PPIIMoL.py` from the repository (see Repository below).
- In PyMOL:
run /full/path/to/PPIIMoL.py
Option B — Clone the repository
git clone https://github.com/silviaenma/PPIIMoL.git
Then in PyMOL:
run PPIIMoL/PPIIMoL.py
Optional: install as a plugin In PyMOL: Plugin → Plugin Manager → Install New Plugin → select `PPIIMoL.py` (or the whole folder) → restart PyMOL.
Usage (GUI)
Once loaded, PPIIMoL opens a Tkinter window with buttons to:
- Load PDB (or use an already-loaded object)
- Detect PPII (scan φ/ψ windows and list segments)
- Scan Cα–H···O=C (optional geometric screening)
- Style / Colors (apply the chosen palette)
- Export (CSV reports; optional per-segment PDBs)
Results are written to a date-stamped folder; selections/objects are created in the PyMOL session and colored according to the chosen scheme.
Example (command line, optional)
# Load the module run /full/path/to/PPIIMoL.py # or run PPIIMoL/PPIIMoL.py # Load a structure and trigger detection fetch 3bog, async=0 ppii_detect()
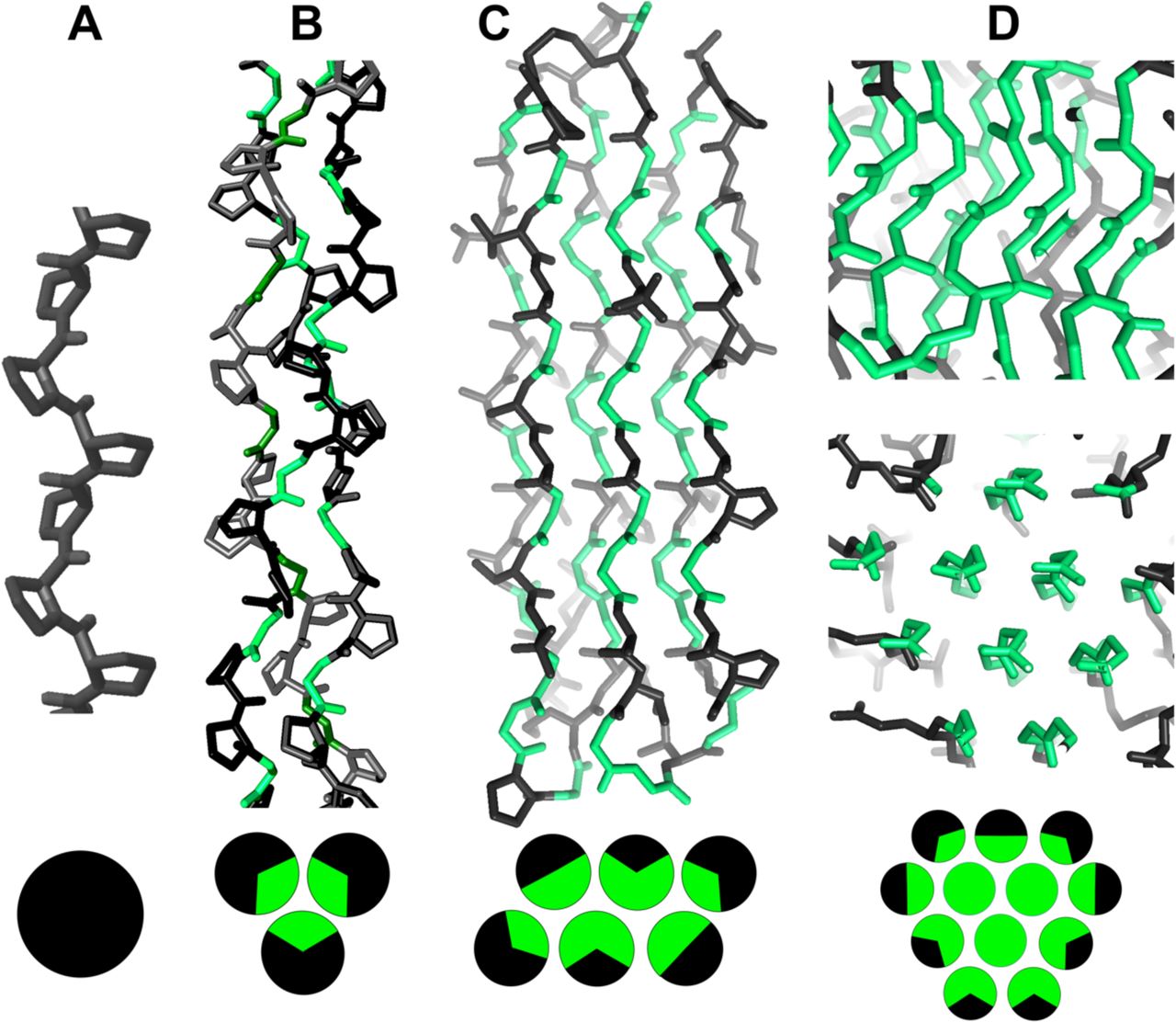
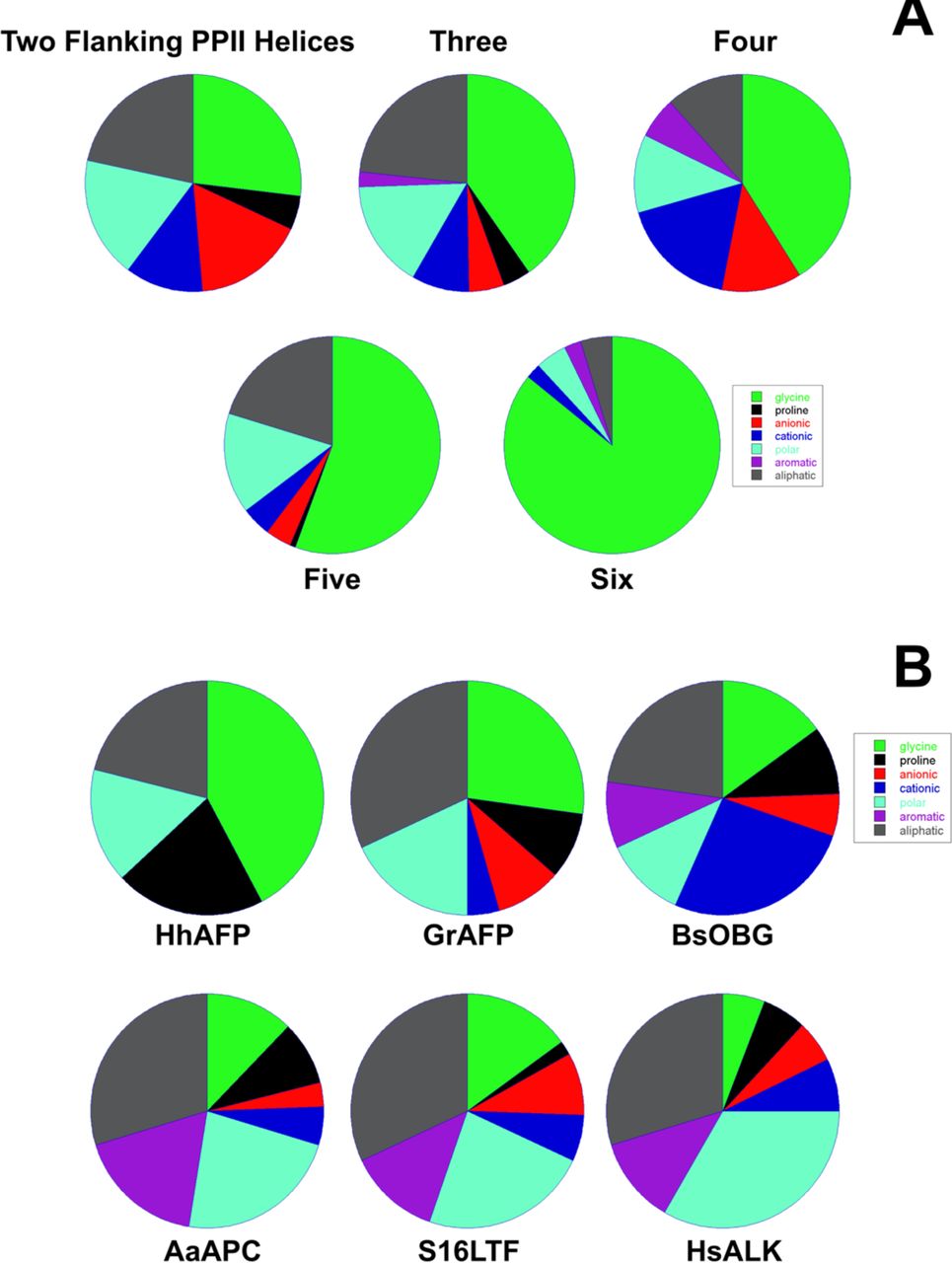
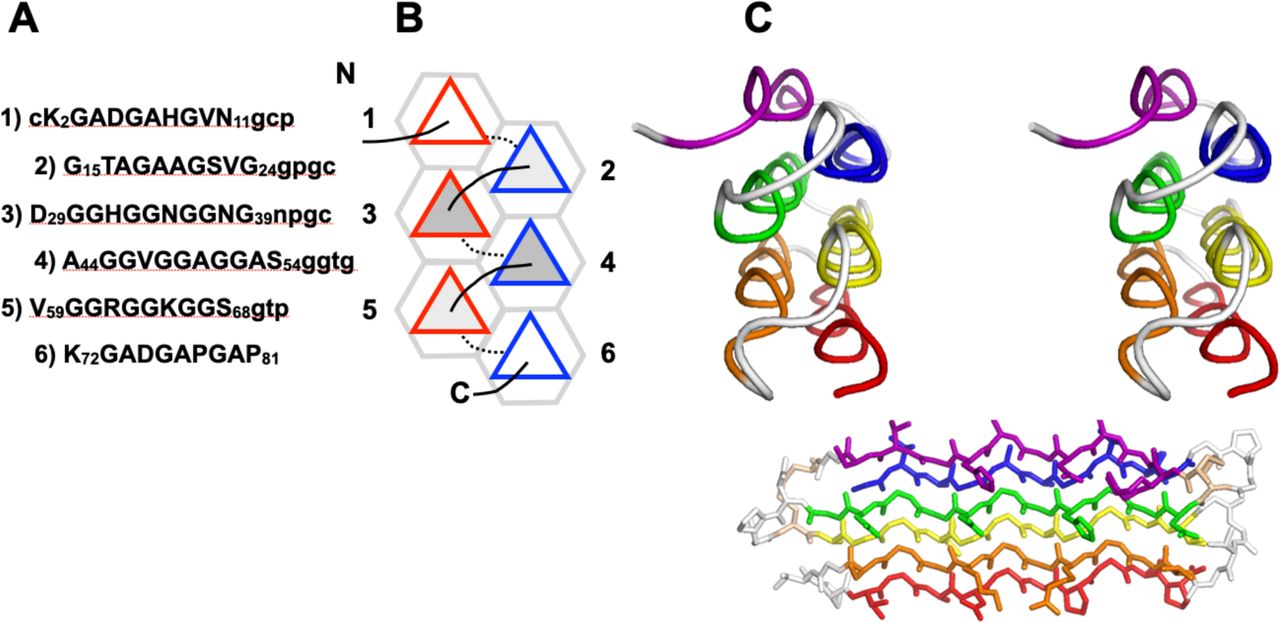
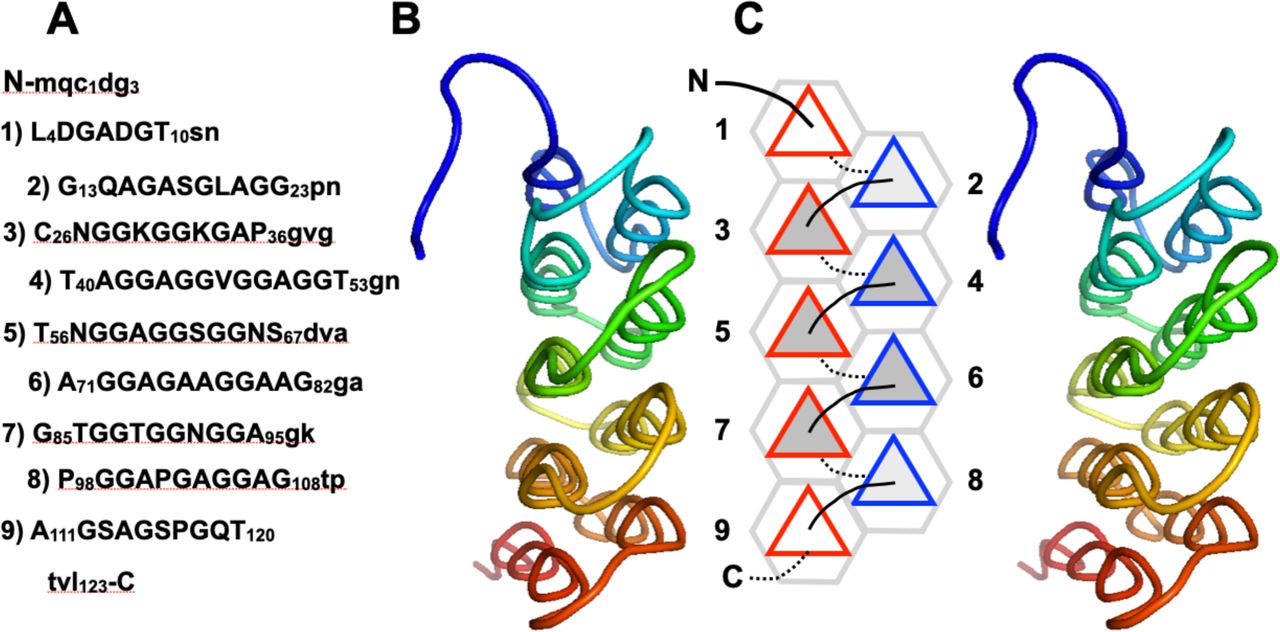
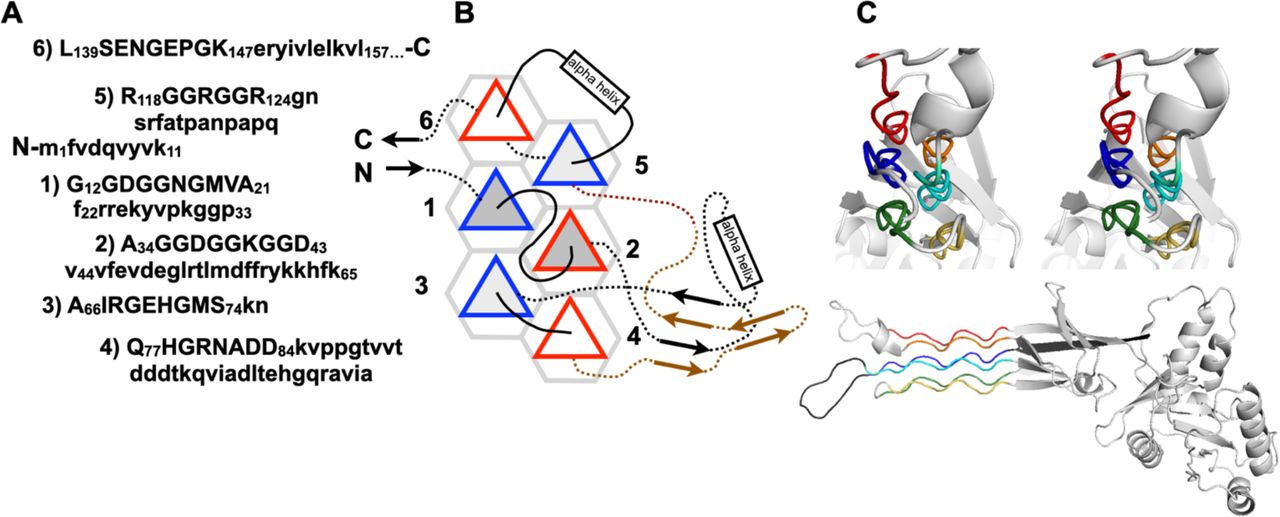
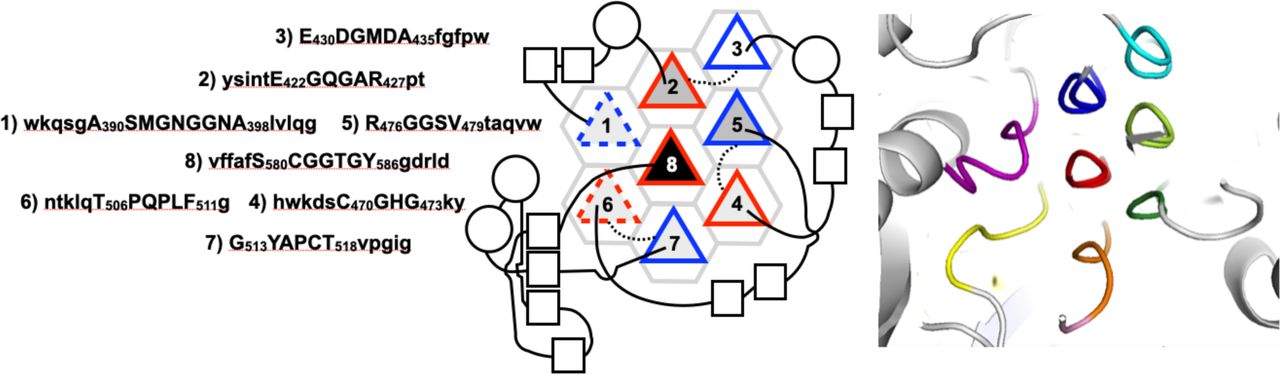
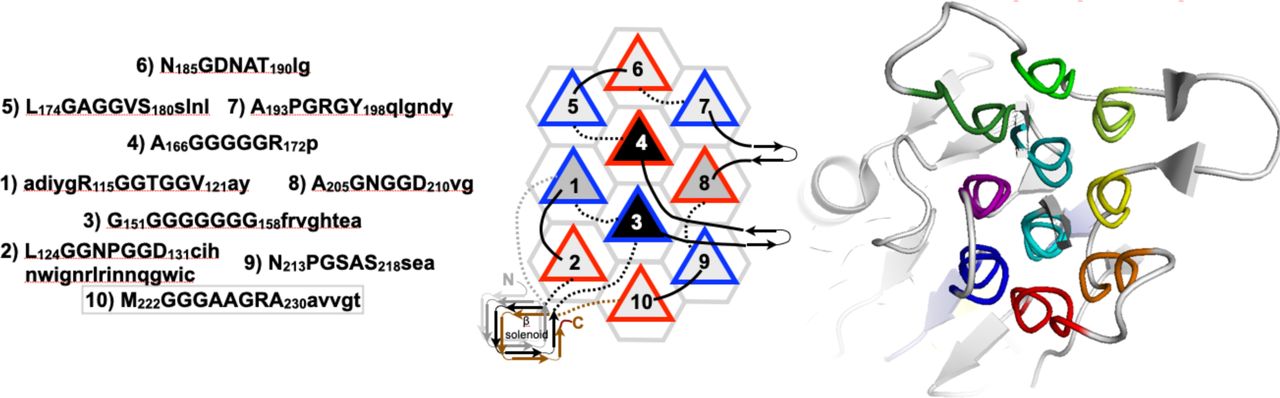
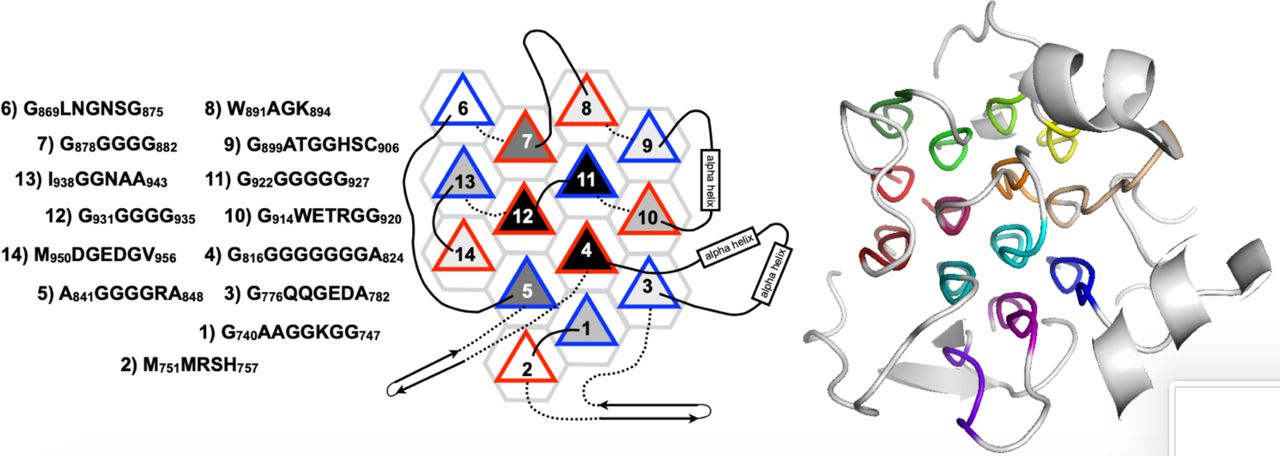
Reference figures
Below are reference figures illustrating PPII bundle organization and residue patterns, reproduced with permission from Segura Rodríguez & Laurents (2024).
GUI buttons
- Load PDB: Opens a file dialog (.pdb/.cif).
- Prepare structure: Optional cleanup (remove solvent/ligands, add hydrogens).
- Detect PPII: Scans backbone torsion angles (φ/ψ), creates selections (ppii_1, ppii_2, …) and colors them.
- Scan Cα–H···O=C: Searches plausible non-canonical contacts; cutoffs configurable.
- Style / Colors: Applies the selected color scheme.
- Export: Writes CSV reports (angles, contacts, detected segments).
Troubleshooting
- GUI does not appear → Ensure PyMOL build includes Tkinter.
- No segments detected → Check backbone completeness; relax φ/ψ windows.
- Too many contacts → Tighten cutoffs in Scan Cα–H···O=C.
- CSV/PDB not written → Verify write permissions.
- Emojis not visible → Cosmetic only.
How to cite
If PPIIMoL is useful in your work, please cite both the software and the reference article:
Software Rodríguez Fernández, S. E. (2025). PPIIMoL (version 1.1) [Computer software]. GitHub. https://github.com/silviaenma/PPIIMoL
Reference article Segura Rodríguez, C. M., & Laurents, D. V. (2024). Architectonic principles of polyproline II helix bundle protein domains. Archives of Biochemistry and Biophysics, 741, 109981. https://doi.org/10.1016/j.abb.2024.109981
Repository
License
PPIIMoL is released under the GNU GPLv3.