3. Joining Atoms
- Step 1
When a molecule is imported into Blender from a PyMOL export (see the first tutorial), it will look like this:
(2,3-BPG from 1B86.pdb)
- Step 2
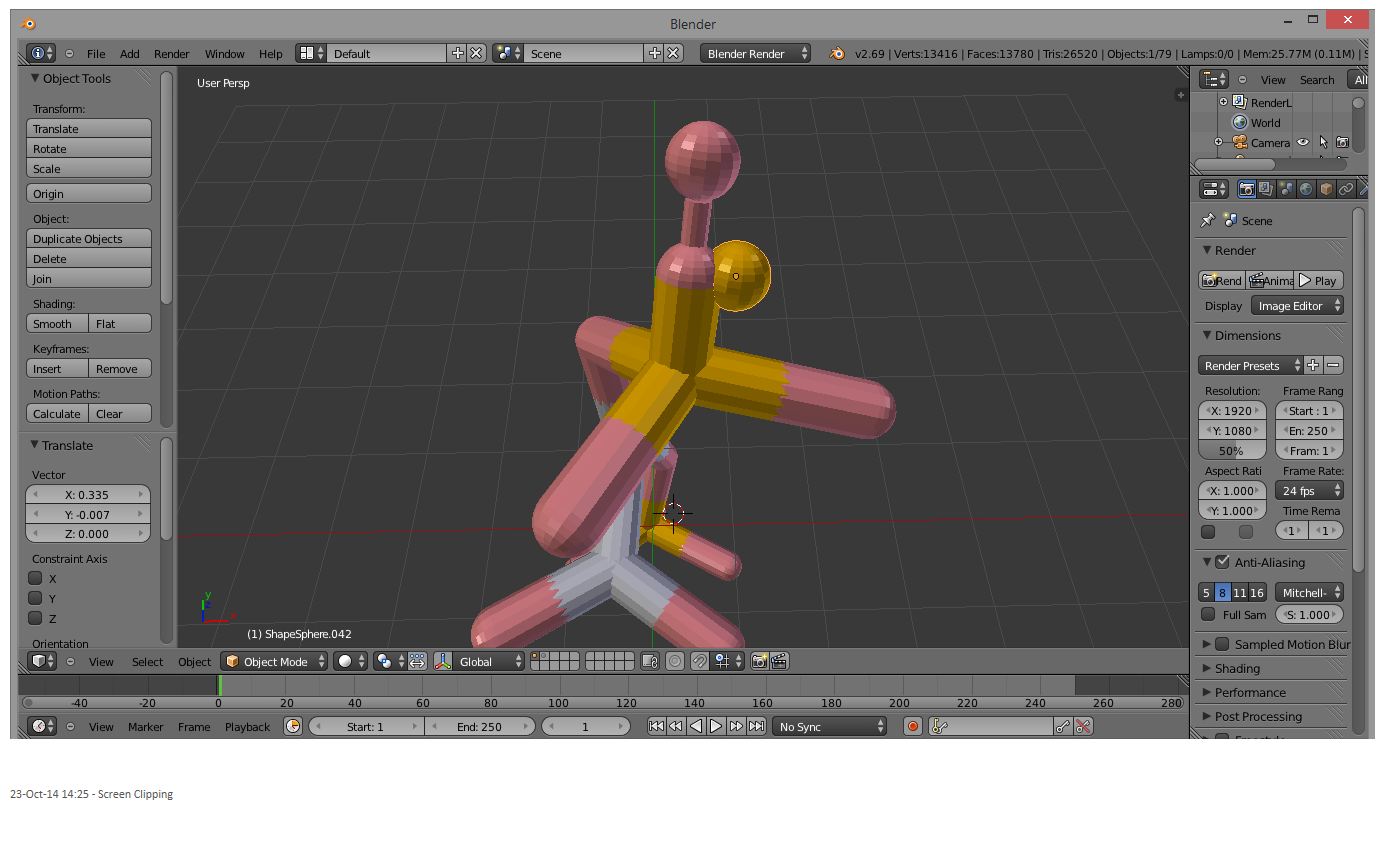
The problem is that this molecule is actually an assembly of spheres and cylinders. When the individual parts are selected using the Shift+right-click method (for scaling, rotating, moving, etc.), some parts are unable to be selected and are left behind. This ruins the molecule:
- Step 3
This problem is solved by grouping the individual pieces. Press b to launch the Blender bounding box tool:
Left-click and drag over the entire molecule. This will select all the pieces:
(selecting)
(all molecule parts selected)
- Step 4
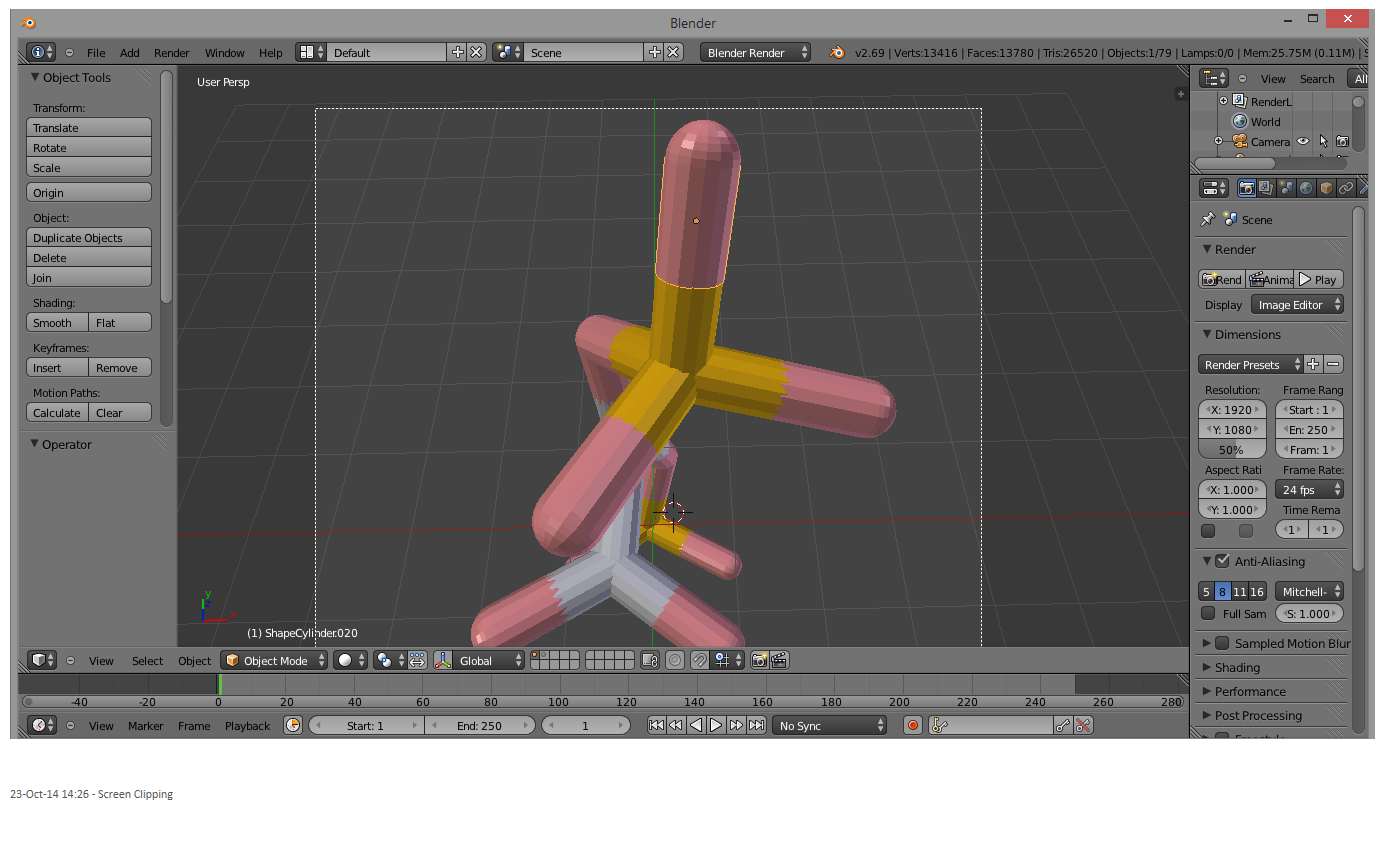
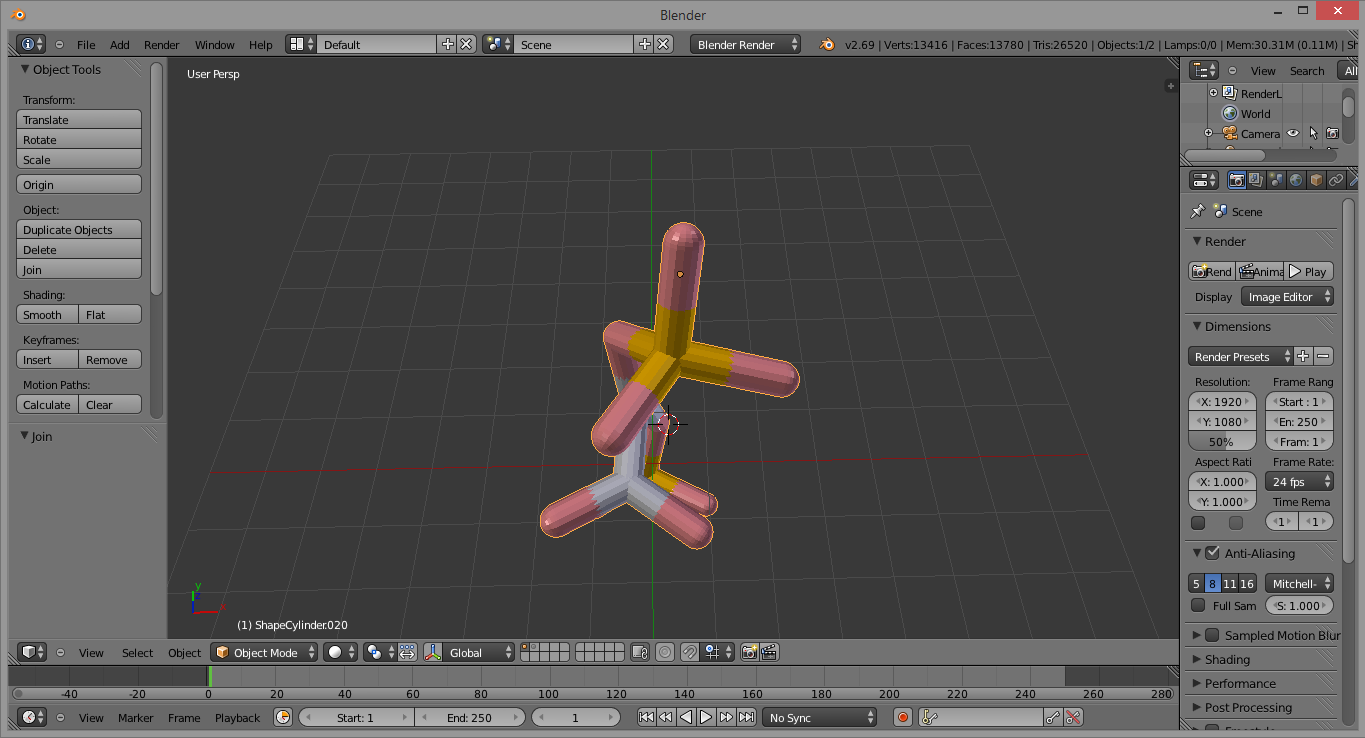
Press Ctrl + j to join all the parts. The final product should look like this:
And there you go! Now the molecule can be moved, rotated, scaled, colored etc. as a whole entity, and selected with a single right-click.
- Step 5
If you are dealing with more than one molecule, you may want to import them into different layers, group them there, and then move them to the main layer.