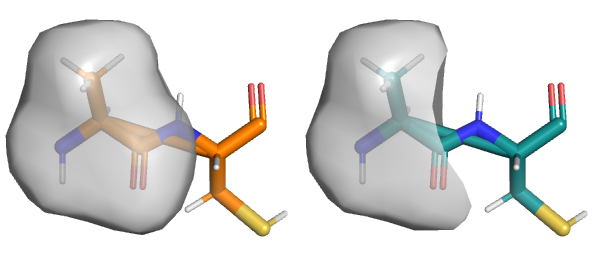Flag
Jump to navigation
Jump to search
Flag is a command to set or clear flags on atom sets. A flag is just some atom-specific property. A flag is either on or off for a residue. Possible flags are:
| Flag name | Value | Description | Notes |
|---|---|---|---|
| focus | 0 | Atoms of Interest (i.e. a ligand in an active site) | Reserved for molecular modeling.
Affects sculpting. |
| free | 1 | Free Atoms (free to move subject to a force-field) | |
| restrain | 2 | Restrained Atoms (typically harmonically constrained) | |
| fix | 3 | Fixed Atoms (no movement allowed) | |
| exclude | 4 | Atoms which should not be part of any simulation | |
| study | 5 | ||
| 6 | Protein (polymer.protein selector)
|
See auto_classify_atoms and auto_show_classified | |
| 7 | Nucleic acid (polymer.nucleic selector)
| ||
| 8-15 | Free for end users to manipulate | ||
| 16-23 | Reserved for external GUIs and linked applications | ||
| exfoliate | 24 | Remove surface from atoms when surfacing (redundant with excluding those atoms from the selection in show surface, sele)
|
Affects surface (with surface_mode=0), dots (with trim_dots=on), get_area |
| ignore | 25 | Ignore atoms altogether when surfacing | |
| no_smooth | 26 | Do not smooth atom position | Affects cartoon |
| 27 | Polymer | See auto_classify_atoms and auto_show_classified
See Selection Algebra "Chemical classes" | |
| 28 | Solvent | ||
| 29 | Organic | ||
| 30 | Inorganic | ||
| 31 | Guide atom (e.g. CA in proteins) |
Usage
flag flag, selection [, action [, quiet ]]
If the auto_indicate_flags setting is true, then PyMOL will automatically create a selection called "indicate" which contains all atoms with that flag after applying the command.
Arguments
- flag = int or str: Flag name or value
- selection = str: atom selection
- action = set|clear|reset: Note that "reset" will set the flag on the given selection, and clear it on all other atoms {default: reset}
Examples
fab AC
# Image on the left
flag ignore, resn CYS
show surface
# Image on the right
flag ignore, all, clear
flag exfoliate, resn CYS
rebuild surface
# in sculpting, ensure the newMethyl group just added doesn't move around
flag fix, newMethyl
# Introspect the flags bitmask
iterate all, print(hex(flags))
# Select atoms with "fix" flag
select fixedatoms, flag 3