States
Jump to navigation
Jump to search
Introduction to States
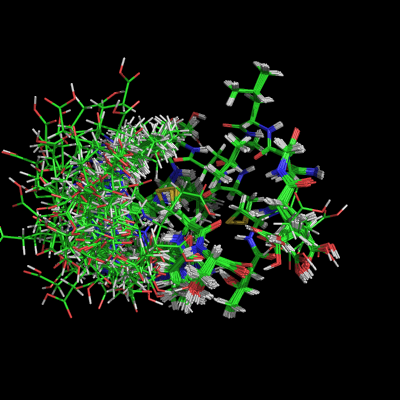
Working with States in PyMOL is a very common task. A State is one particular conformation of an object. For example, one could load an NMR ensemble and set all_states or use the command Split_States to see all entries in the NMR file. Another example could be the set of states from a molecular dynamic (MD) simulation. Each conformer in the MD ensemble could be viewed as a state in PyMOL.
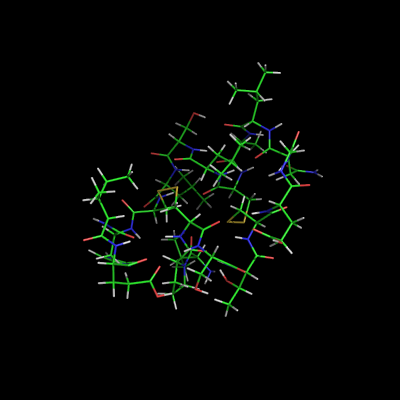
If you are making a movie of a static coordinate set (such as a single crystal structure) then you have only one state. All objects in PyMOL can potentially consist of multiple states. For movies, see Frames.
Using States in PyMOL
- Upon loading an object you can separate each state into its own object with Split_States.
- States can be colored individually.
- One can show all states with the all_states setting.
- One can iterate over states using Iterate_State or change properties with states using the Alter_State function.