PPIIMoL
PPIIMoL
PPIIMoL is a Python module for PyMOL that automates the detection of polyproline II (PPII) helices in proteins. It identifies PPII-like φ/ψ angle patterns, screens for plausible non‑canonical Cα–H···O=C contacts, and provides one‑click visualization and export.
This tool was developed as part of a Bachelor's Thesis in Computer Engineering in collaboration with the Protein Structure, Dynamics and Interactions by NMR Group at the Instituto de Química‑Física “Blas Cabrera” (IQF‑CSIC). The module’s design and validation take as a primary reference the data and architectonic principles reported by Segura Rodríguez & Laurents (2024). See Provenance & how to cite below.
Scientific background
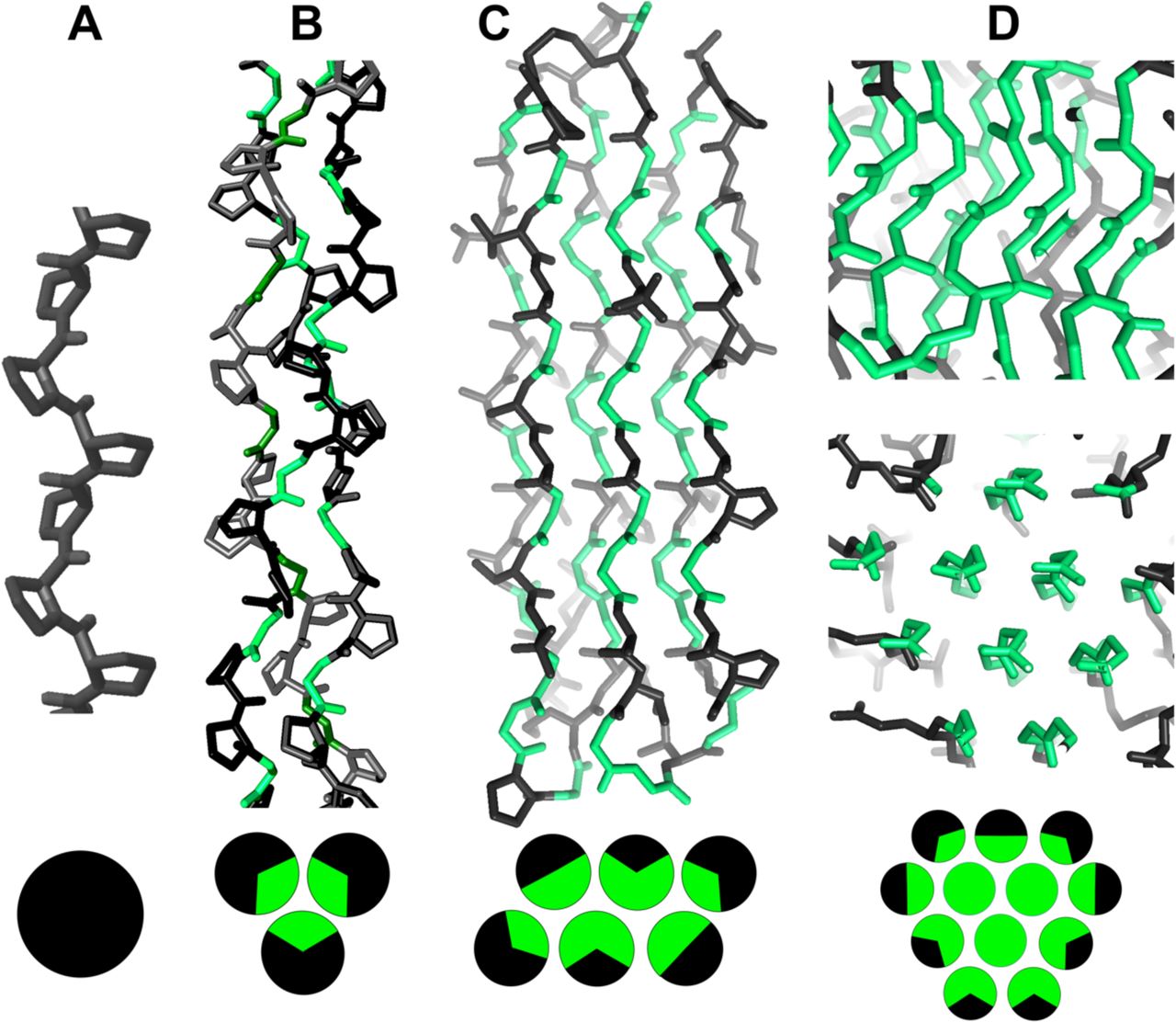
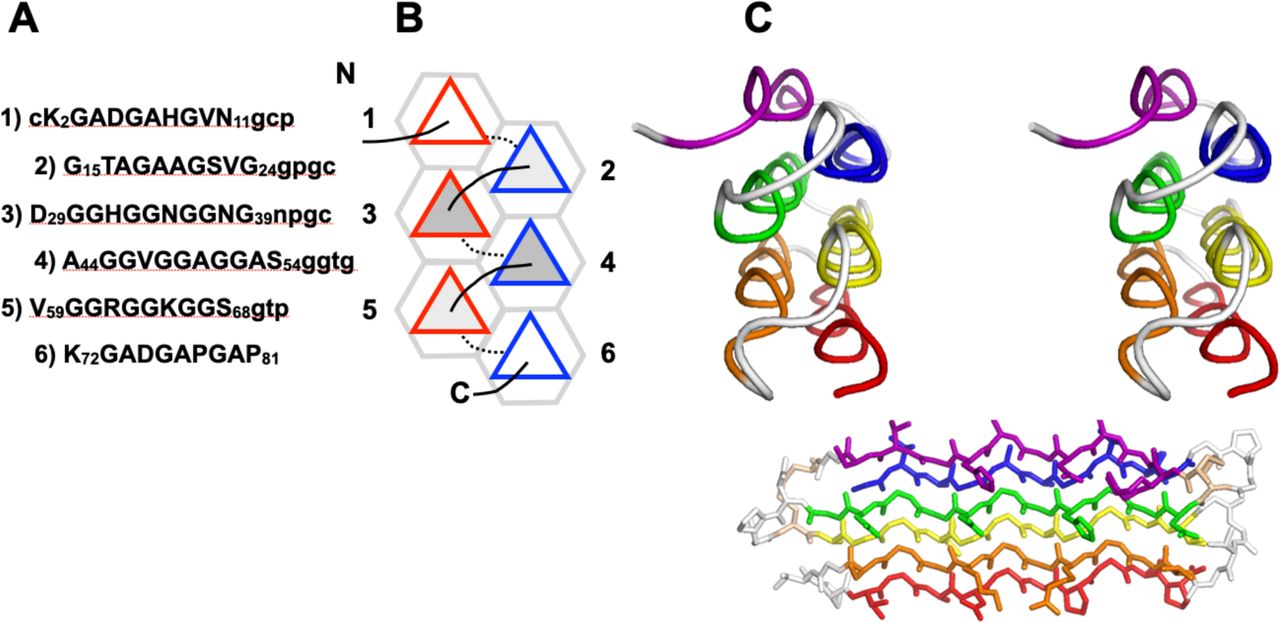
Polyproline II (PPII) helices are extended, left‑handed motifs (≈3 residues/turn) typically enriched in glycine‑ and proline‑rich domains. Although common in several glycine‑rich bundles, they are often unannotated in PDB files. PPIIMoL automates their detection directly in PyMOL to improve speed and reproducibility.
Features
- 🔍 Automatic detection of PPII segments via phi/psi angle analysis.
- 🧬 Identification of Cα-H···O=C interactions relevant to structural stability.
- 📊 CSV export of detected segments and interactions.
- 🎨 Direct visualization in PyMOL with customizable color codes.
- 🖱️ Simple Tkinter-based GUI — no commands required; all actions are accessible via buttons.
Requirements
Installation
Option A — Single‑file run (no cloning)
- Download `PPIIMoL.py` from the GitHub repository.
- In PyMOL:
run /path/to/PPIIMoL.py
Option B — Clone repository (optional)
git clone https://github.com/silviaenma/PPIIMoL.git
Then in PyMOL:
run PPIIMoL/PPIIMoL.py
Option C — Install as plugin (optional)
- PyMOL → Plugin → Plugin Manager → Install New Plugin → select `PPIIMoL.py` (or the folder) → restart.
Usage
When loaded, PPIIMoL opens a Tkinter window with buttons to:
- Prepare the structure (remove solvent, add hydrogens)
- Detect PPII segments
- Visualize / color segments
- Export results (CSV, PDB)
Outputs are written to a dated folder and objects/selections are created in the PyMOL session.
Figures
Provenance & how to cite
The algorithm implements the manual workflow used at IQF‑CSIC and was benchmarked on reference domains reported by Segura Rodríguez & Laurents. If you use PPIIMoL in research, please cite:
- Software — Rodríguez Fernández, S. E. PPIIMoL: automated detection of polyproline II helices in PyMOL. GitHub: https://github.com/silviaenma/PPIIMoL (GPL‑3.0).
- Primary reference (journal) — Segura Rodríguez, C. M., & Laurents, D. V. (2024). Architectonic principles of polyproline II helix bundle protein domains. Archives of Biochemistry and Biophysics, 741, 109981. DOI
- Figures source (preprint license) — bioRxiv preprint of the above article, licensed under CC BY‑NC‑ND 4.0 (attribution required; no derivatives).
Repository
License
PPIIMoL is released under the GNU GPLv3.