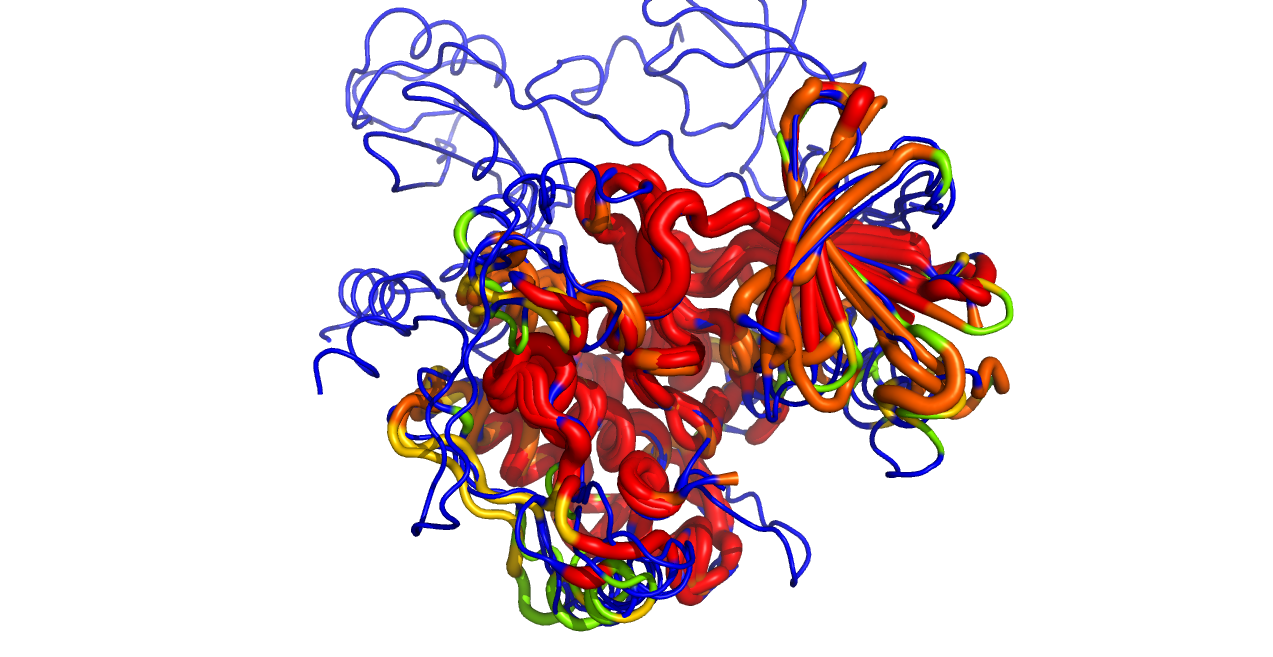
Color by conservation
Jump to navigation
Jump to search
Overview
This script reads an alignment object and colors the protein objects in the alignment by the sequence conservation found in the alignment.
Example Usage
# run the script
run show_conserved.py
# turn on the sequence viewer
set seq_view
# get some kinases
fetch 1opk 3dtc 3p86 2eva 3efw, async=0
# align them into the "aln" object
for x in cmd.get_names(): cmd.align(x, "3efw and c. A", object="aln")
# color
color_by_conservation("aln", as_putty=1)
The Code
def color_by_conservation(aln, names=(), color="rainbow", as_putty=0, _self=cmd):
"""
PARAMETERS
aln
(string) the alignment object name
names
(list) a list of object names that are in the alignment;
if (), then PyMOL will attempt to glean the names
from the alignment object
color
(string) valid PyMOL spectrum name
as_putty
(0 or 1) if 0 display is not changed, else participating objects are shown
as cartoon putty, colored by the 'color' field
"""
# PyMOL doesn't yet know about object:alignment
# but we need to check that this exists or we might crash
if _self.get_type(aln)!="object:":
print "Error: Bad or incorrectly specified alignment object."
return None
r = cmd.get_raw_alignment(aln)
if names==():
known_objs = []
map(known_objs.extend, map(lambda x: map(lambda y: y[0], x), r))
known_objs=set(known_objs)
# highest number of matches seen
M = max(map(len,r)) + 1
else:
known_objs = set(names)
M = len(known_objs) + 1
for obj in known_objs:
_self.alter(obj, "b=0.0")
for af in r:
c = float(1.0 + len(af)) / float(M)
for y in af:
_self.alter("%s and index %s" % (y[0], y[1]), "b=c", space={'c':c})
if as_putty!=0:
for obj in known_objs:
_self.show_as("cartoon", "%s" % obj)
_self.cartoon("putty", "%s" % obj)
_self.spectrum('b', color, obj)
_self.sort()
_self.rebuild()
return None