Ccp4 ncont: Difference between revisions
No edit summary |
|||
| Line 26: | Line 26: | ||
== The Code == | == The Code == | ||
<source lang="python"> | <source lang="python"> | ||
import re | |||
def parseContacts( f ): | |||
# /1/B/ 282(PHE). / CE1[ C]: /1/E/ 706(GLN). / O [ O]: 3.32 | # /1/B/ 282(PHE). / CE1[ C]: /1/E/ 706(GLN). / O [ O]: 3.32 | ||
# conParser = re.compile("\s*/(\d+)/([A-Z])/\s*(\d+).*?/\s*([A-Z0-9]*).*?:") | # conParser = re.compile("\s*/(\d+)/([A-Z])/\s*(\d+).*?/\s*([A-Z0-9]*).*?:") | ||
| Line 55: | Line 55: | ||
print "Unknown mode", mode | print "Unknown mode", mode | ||
def selectContacts( contactsfile, selName1 = "source", selName2 = "target" ): | |||
""" | """ | ||
selectContacts -- parses CCP4 NCONT log file and selects residues and atoms from the list of the contacts found. | selectContacts -- parses CCP4 NCONT log file and selects residues and atoms from the list of the contacts found. | ||
| Line 98: | Line 98: | ||
cmd.select( atomName, atomName + " or " + chain+"/"+residue+"/"+atom) | cmd.select( atomName, atomName + " or " + chain+"/"+residue+"/"+atom) | ||
cmd.extend("selectContacts", selectContacts) | |||
</source> | </source> | ||
[[Category:Script_Library]] [[Category:ThirdParty Scripts]] [[Category:Structural Biology Scripts]] | [[Category:Script_Library]] [[Category:ThirdParty Scripts]] [[Category:Structural Biology Scripts]] | ||
Revision as of 08:49, 26 May 2011
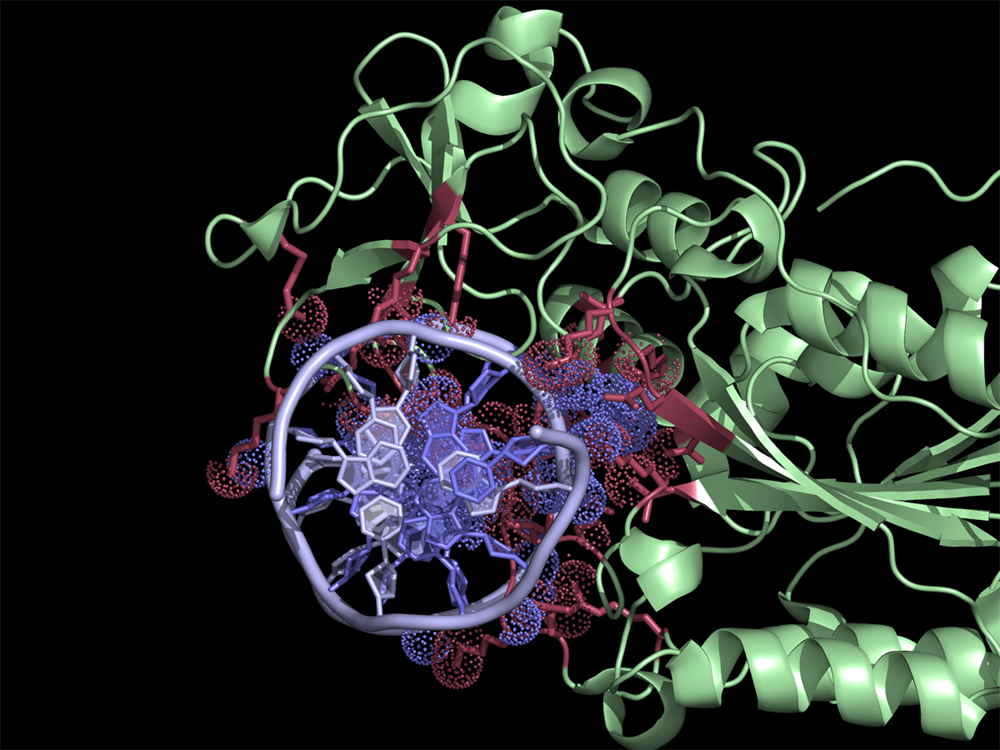
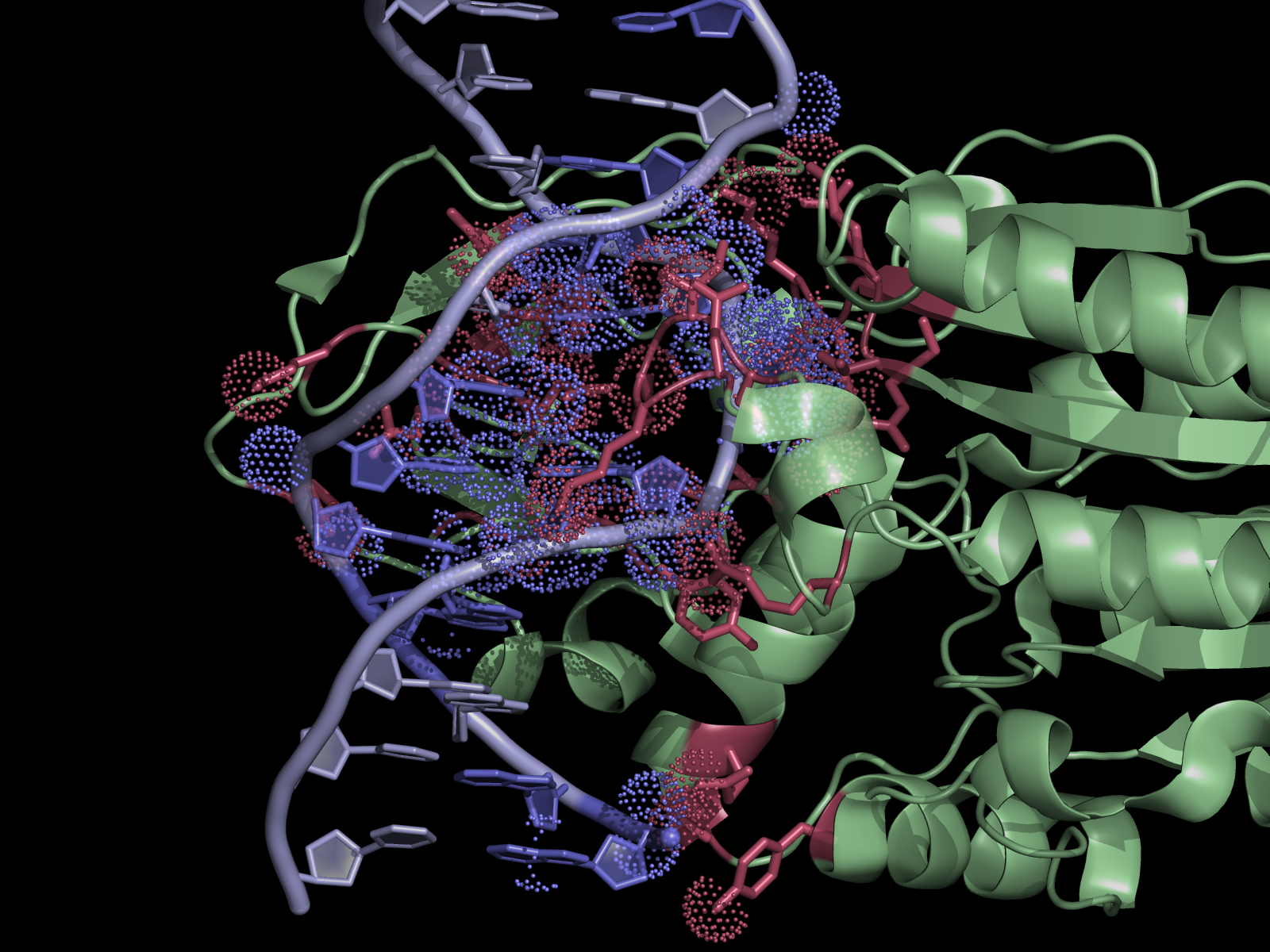
Overview
The script selects residues and atoms from the list of the contacts found by NCONT from CCP4 Program Suite (NCONT analyses contacts between subsets of atoms in a PDB file). First, we run NCONT on our pdb file to find interface residues. Then by using the ContactsNCONT script in PyMOL we separately select residues and atoms listed in a ncont.log file. This generates two selections (atoms and residues) for each interacting chain, allowing quick manipulation of (sometimes) extensive lists in NCONT log file.
This script works best for intermolecular contacts (when NCONT target and source selections don't overlap). If crystal contacts (NCONT parameter cell = 1 or 2) are included then additional coding is required to distinguish inter from intramolecular contacts.
Usage
selectContacts( contactsfile, selName1 = "source", selName2 = "target" )
Examples
First use NCONT to find interface residues/atoms in the pdb file. Once you have ncont.log file proceed to PyMOL. Make sure you've run the ContactsNCONT script first.
fetch 2c7r selectContacts ncont.log, selName1=prot, selName2=dna
The Code
import re
def parseContacts( f ):
# /1/B/ 282(PHE). / CE1[ C]: /1/E/ 706(GLN). / O [ O]: 3.32
# conParser = re.compile("\s*/(\d+)/([A-Z])/\s*(\d+).*?/\s*([A-Z0-9]*).*?:")
conParser = re.compile("\s*/(\d+)/([A-Z]*)/\s*(\d+).*?/\s*([A-Z0-9]*).*?:") # * is needed when chain code is blank
mode = 0
s1 = []
s2 = []
pairs = []
for line in f:
if mode == 0:
if line.strip().startswith("SOURCE ATOMS"):
mode = 1
elif mode == 1:
mode = 2
elif mode == 2:
matches = conParser.findall(line)
if len(matches) == 0:
return (s1, s2, pairs)
if len(matches) == 2:
s1.append(matches[0])
s2.append(matches[1])
elif len(matches) == 1:
s2.append(matches[0])
pairs.append((len(s1)-1, len(s2)-1))
else:
print "Unknown mode", mode
def selectContacts( contactsfile, selName1 = "source", selName2 = "target" ):
"""
selectContacts -- parses CCP4 NCONT log file and selects residues and atoms from the list of the contacts found.
PARAMS
contactsfile
filename of the CCP4 NCONT contacts log file
selName1
the name prefix for the _res and _atom selections returned for the
source set of chain
selName2
the name prefix for the _res and _atom selections returned for the
target set of chain
RETURNS
* 2 selections of interface residues and atoms for each chain are created and named
depending on what you passed into selName1 and selName2
AUTHOR:
Gerhard Reitmayr and Dalia Daujotyte, 2009.
"""
# read and parse contacts file into two lists of contact atoms and contact pair list
s1, s2, pairs = parseContacts(open(contactsfile))
# create a selection for the first contact list
resName = selName1 + "_res"
atomName = selName1 + "_atom"
cmd.select(resName, None)
cmd.select(atomName, None)
for (thing, chain, residue, atom) in s1:
cmd.select( resName, resName + " or " + chain+"/"+residue+"/")
cmd.select( atomName, atomName + " or " + chain+"/"+residue+"/"+atom)
# create a selection for the second contact list
resName = selName2 + "_res"
atomName = selName2 + "_atom"
cmd.select(resName, None)
cmd.select(atomName, None)
for (thing, chain, residue, atom) in s2:
cmd.select( resName, resName + " or " + chain+"/"+residue+"/")
cmd.select( atomName, atomName + " or " + chain+"/"+residue+"/"+atom)
cmd.extend("selectContacts", selectContacts)