Ramp New: Difference between revisions
No edit summary |
|||
| Line 8: | Line 8: | ||
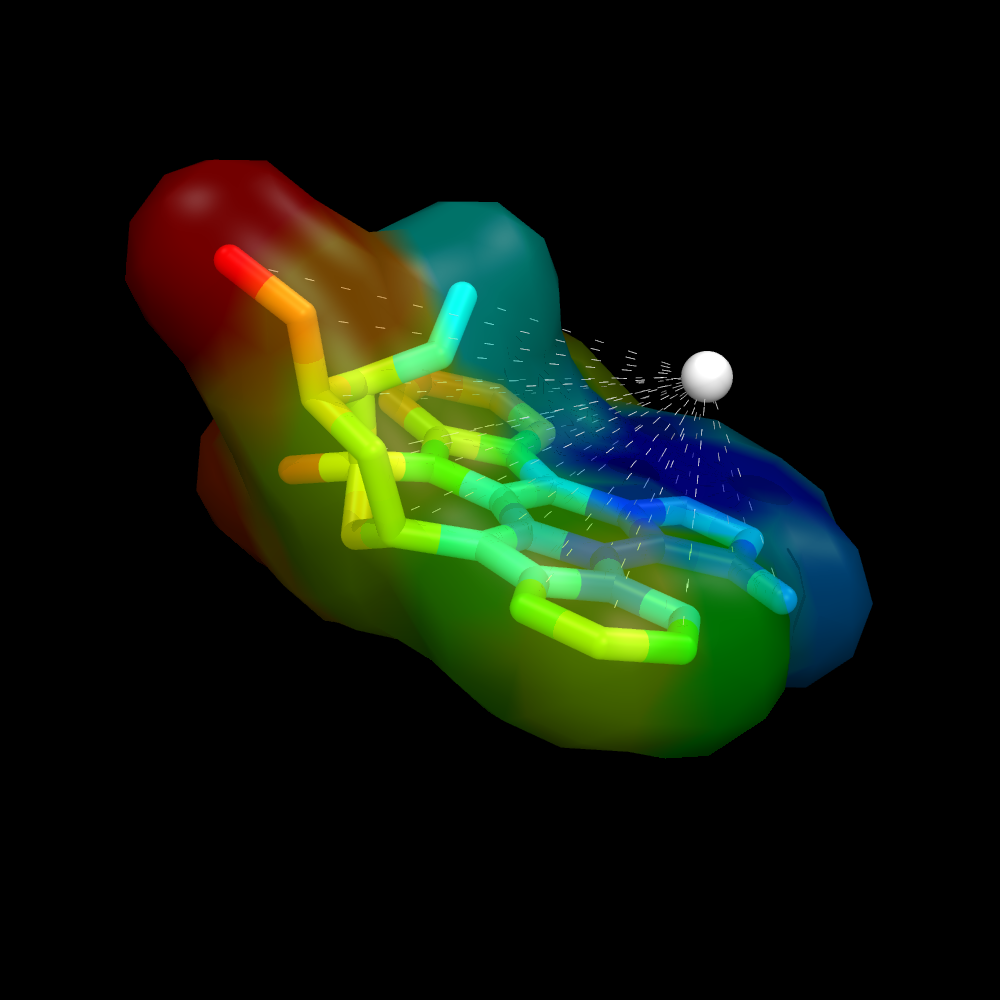
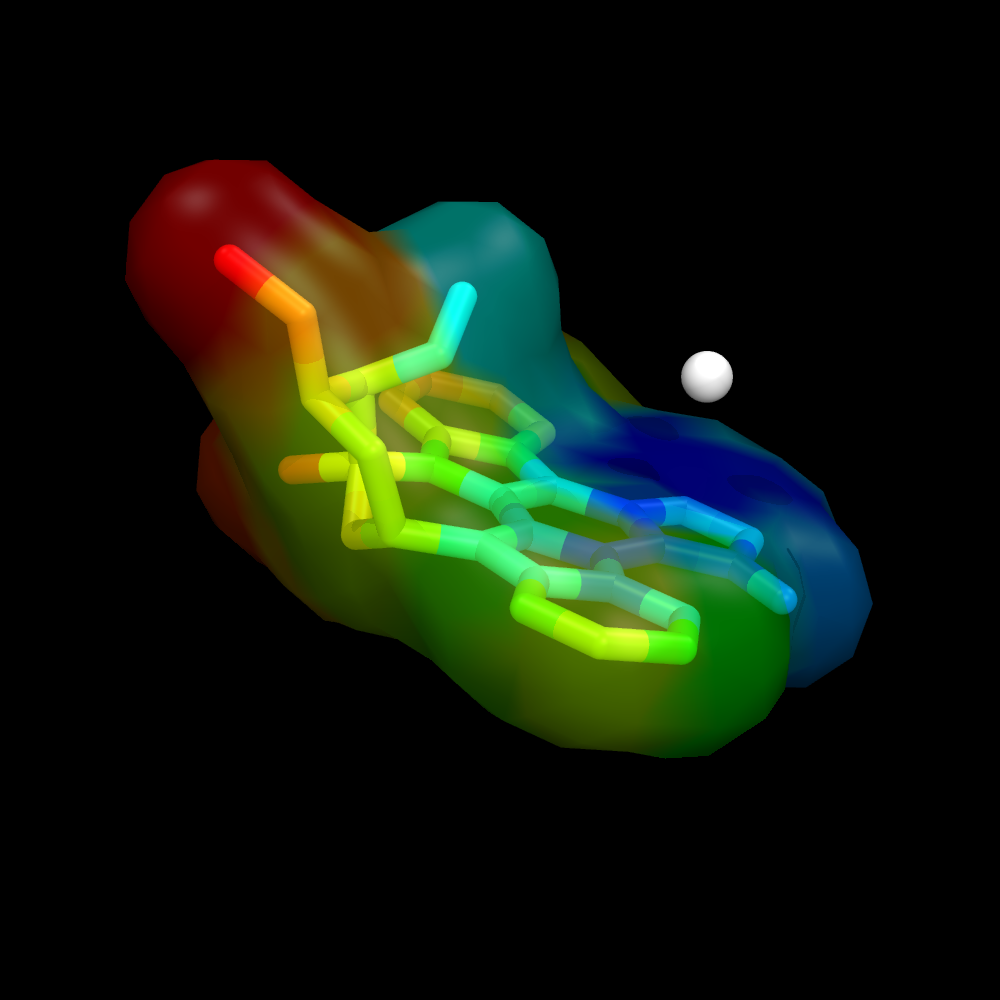
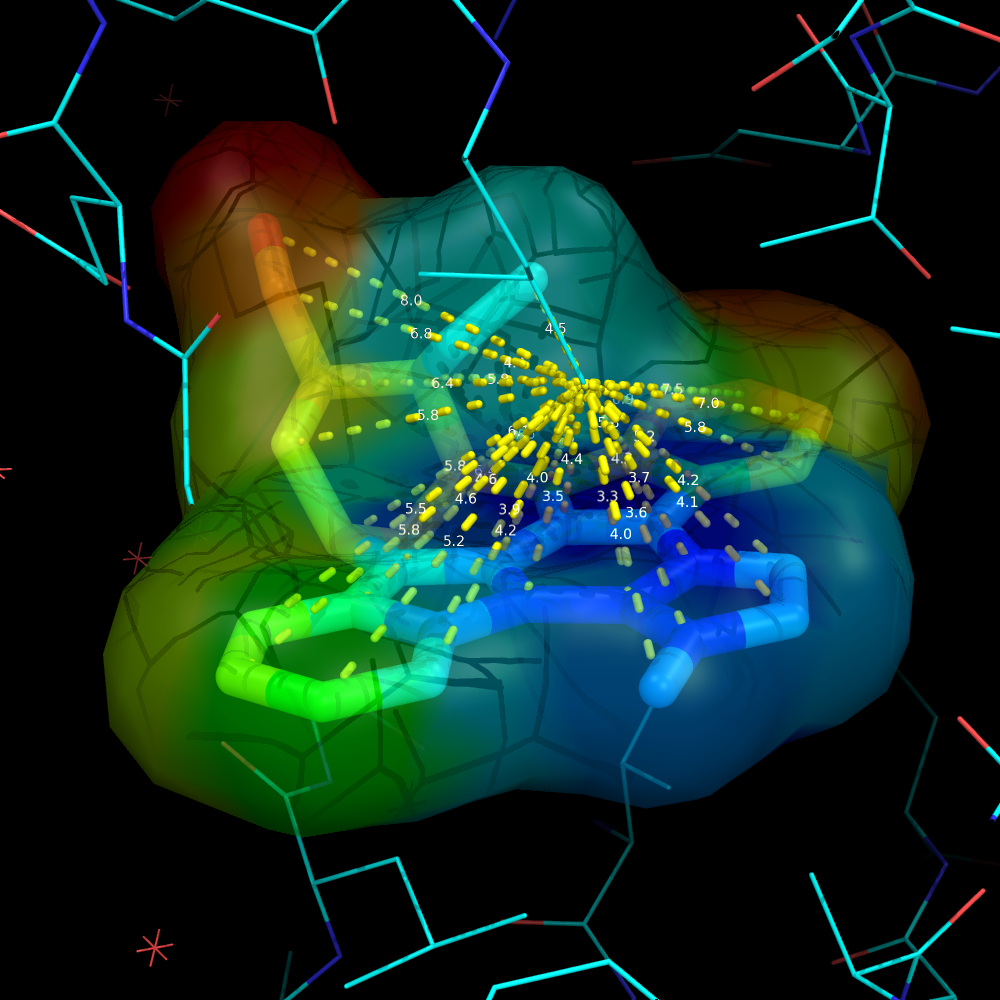
Using a ramp and a distance measure, we can color the surface by some property--here, I'll chose distance from some important atom in the receptor to the ligand atom. | Using a ramp and a distance measure, we can color the surface by some property--here, I'll chose distance from some important atom in the receptor to the ligand atom. | ||
<gallery perrow=3 widths= | <gallery perrow=3 widths=200 heights=200> | ||
Image:Surface_by_prop3.png|Ligand surface colored by distance from some given atom. The remainder of the protein is hidden to more clearly visualize the calculated distances and surface color | Image:Surface_by_prop3.png|Ligand surface colored by distance from some given atom. The remainder of the protein is hidden to more clearly visualize the calculated distances and surface color | ||
Image:Surface_by_prop2.png | Image:Surface_by_prop2.png | ||
Revision as of 16:20, 4 July 2010
ramp_new creates a color ramp based on a map potential value or based on proximity to a molecular object.
Color ramps are extremely powerful but complicated to use.
In the simplest case, they can be used to color representations based on the potential values found in a map object at the corresponding positions in space. In another simple case, representations can be colored based on proximity to a target. Note that since ramp targets must themselves be real objects (not merely selections), the "create" command may be needed in order to generate an appropriate target. In more complicated cases, they can be used to color representations on one object based atoms found in another. Ramps can operate recursively. In other words, the output color from one ramp can be used as the input color for another. For example, you could color by map potential within a certain distance of the target object, beyond which, a uniform color is applied.
Example
Using a ramp and a distance measure, we can color the surface by some property--here, I'll chose distance from some important atom in the receptor to the ligand atom.
To reproduce the results shown here you must do the following:
- obtain a protein
- calculate some property for some set of atoms (like distance from some central location) and stuff the values into the b-factor
- create a new object from the atoms for which you just measured a property
- create a new ramp from the object with ramp_new
- set the surface color of the new object
Usage
# normal PyMOL usage
ramp_new name, map_name [, range [, color [, state [, selection [,
beyond [, within [, sigma [, zero ]]]]]]]]
# in the API
cmd.ramp_new(string name, string map_name, list range, list color,
int state, string selection, float beyond, float
within, float sigma, int zero, int quiet)
- name = string: name of the ramp object
- map_name = string: name of the map (for potential) or molecular object (for proximity)
- range = list: values corresponding to slots in the ramp
- color = list: colors corresponding to slots in the ramp
- state = integer: state identifier
- selection = selection: for automatic ranging
- beyond = number: with automatic ranging, are we excluding values beyond a certain distance from the selection?
- within = number: with automatic ranging, are we only including valuess within a certain distance from the selection?
- sigma = number: with automatic ranging, how many standard deviations from the mean do we go?
- zero = integer: with automatic ranging, do we force the central value to be zero?
Example
Simple Example
ramp_new e_pot_color, e_pot_map, [-10,0,10], [red,white,blue]
Normal Usage
load pdbset1.pqr
load pdbset_1.dx
load pdbset3.pqr
load pdbset_3.dx
ramp_new ramp1, pdbset_1, [ -3, 0, 3]
ramp_new ramp3, pdbset_3, [ -3, 0, 3]
show surface
set surface_ramp_above_mode
set surface_color, ramp1, pdbset1
set surface_color, ramp3, pdbset3