MAC Install: Difference between revisions
(→Gratuitous Eye-Candy Tweaks: This no longer works for reasons obscure to me.) |
|||
| Line 110: | Line 110: | ||
---- | ---- | ||
===PyMOLX11Hybrid=== | ===PyMOLX11Hybrid=== | ||
Revision as of 22:45, 21 November 2007
http://images.apple.com/powermac/images/solutionsscience20050427.jpg
Installing MacPyMOL
http://delsci.com/macpymol/macpymol350.jpg
Essentials
The download is about as straightforward as it gets, and you can install it wherever it makes you happy. You need a 3 button mouse (clickable scroll wheel = middle button). Apple has finally come to its senses and designed a proper, ergonomically pleasant, scrollbutton mouse that works well with pymol and permits horizontal scrolling. Most other mice will also work well.
Warning on Mouse Drivers
One word of warning: Do not install 3rd party drivers for multi-button mice if you can avoid it. These often mess with the mapping of the middle button or have other horrific side effects. Fortunately, with OS X, you should not need to.
Invoking pymol from the unix command line
The unix executable resides at MacPyMOL.app/Contents/MacOS/MacPyMOL
I (Bill Scott) wrote a cheezy pymol shell script (and zsh function) to invoke this on the command line. It uses mdfind to find the executable. I also use this ~/.pymolrc file.
Additional invokation options and further details are discussed under Launching_PyMOL#MacOS_X:
Extras
You don't need any of these to use MacPyMOL. But you didn't really need a Mac either. Face it: You need these.
Mighty Mouse
http://images.apple.com/mightymouse/images/index360scroll220050802.gif
A 3-button mouse is essential. Apple's Mighty Mouse is an extra treat.
PowerMate Dial
http://www.griffintechnology.com/assets/images/products/powermate/prod_powermate_sub01b.jpg
The PowerMate dial works nicely with pymol.
Stereo
http://images.apple.com/powermac/images/graphicspymol20051018.jpg
The latest Macs finally support stereo in a window. There is more information in the Monitors Hardware Options section.
Second Monitor
The trick to getting MacPyMOL to work in stereo on the second monitor is to force it to initially open on that display by providing an appropriate "-X #" (and perhaps -Y #) option on launch. That way the OpenGL context will be created with stereo support.
./MacPyMOL.app/Contents/MacOS/MacPyMOL -X -1000
./MacPyMOL.app/Contents/MacOS/MacPyMOL -X -1000 -Y 100
Source: Warren DeLano; PyMOL Users Archive
Installing X-windows based pymol on Mac OS X
Why would you want to do this?
- You want to run a free, guilt-free, open-source version of pymol
- You just happen to prefer the tkinter menu
- You want to use plugins, for example, the apbs plugin for free grasp-like electrostatic calculations.
Simplest Installation
Install pymol with Fink
http://pdb.finkproject.org/img/mlogo.png
By far the simplest way to install the X-windows based version of pymol on OS X is by using the fink package management system. To compile it, all you need to do is issue the command
fink install pymol-py25
(This will install python2.5 in fink, along with an X-windows based tkinter. There are also versions that permit you to install pymol to interact with python2.4 and even python2.3. Fink uses its own unix-type python installation.)
The fink pymol package currently exists in the unstable branch of fink, so you will either have to activate the unstable branch or make the following symbolic links:
sudo ln -s /sw/fink/dists/unstable/main/finkinfo/sci/pymol-py.* /sw/fink/dists/local/main/finkinfo/.
You might need to create the latter directory if it doesn't already exist, i.e., issue the command
sudo mkdir -p /sw/fink/dists/local/main/finkinfo
10.4 ONLY: Be sure to set your display environment in your start up shell script to use pymol. For example in your home directory, the .bashrc (or .bash_profile) file should contain:
export DISPLAY=":0.0"
Do NOT set the DISPLAY variable for 10.5, as God does this for you automatically.
I (wgscott) have put a whole lot of further information on how to use fink to install crystallographic software on my own wiki and website, including instructions on how to install precompiled binary packages using fink.
The fink pymol package is currently maintained by Jack Howarth.
Install APBS and friends with fink
To use the electrostatics plugin, you will need APBS and its dependencies. These are also available as fink packages, and include apbs, maloc and pdb2pqr. If you have multiple processors available, you might wish to install the mpi version of apbs.
Issuing the command
fink install apbs
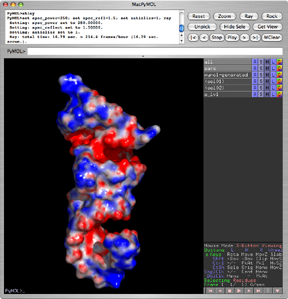
will install apbs and its required dependencies for you. The fink pymol package is already preconfigured to do the right thing to use apbs as a plugin. Here is a big screenshot of the fink APBS package being invoked via the pymol plugin.
Nucleic acids may prove problematic for the apbs plugin. If so, use the pdb2pqr command-line tool to create a pqr file manually, instead of using the plugin to generate it.
PyMOLX11Hybrid
MacPyMOL for Tiger now includes a hybrid X11 mode. Assuming that X11 is already installed, simply duplicate MacPyMOL.app and rename it "PyMOLX11Hybrid.app" and launch.
Doing this creates the inverse situation of the previous tweak; the molecular viewer window is aqua-based and the tkinter GUI is X11 based. (Naturally, I had to try the trick of making an aqua-tkinter GUI. It sort of works, but the GUI is dead, thanks to a threading bug in the aqua Tcl/Tk framework that comes with Apple. Bummer.)