APBS Electrostatics Plugin: Difference between revisions
(created) |
(Python API) |
||
| Line 117: | Line 117: | ||
* Calculate Map with APBS > Options >> Grid Spacing: 2.0 | * Calculate Map with APBS > Options >> Grid Spacing: 2.0 | ||
* click "Run" ''(takes about 20 minutes!)'' | * click "Run" ''(takes about 20 minutes!)'' | ||
== Python API == | |||
''Disclaimer: The plugin code is not part of the official '''cmd''' API and could be subject to change.'' | |||
Example with PDB [https://www.rcsb.org/structure/1ubq 1ubq]: | |||
<syntaxhighlight lang="python"> | |||
from pmg_tk.startup.apbs_gui.creating import pdb2pqr_cli | |||
from pmg_tk.startup.apbs_gui.electrostatics import map_new_apbs | |||
cmd.fetch("1ubq") | |||
pdb2pqr_cli("prepared01", "1ubq", options=["--ff", "amber"]) | |||
map_new_apbs("apbs_map01", "prepared01") | |||
cmd.ramp_new("apbs_ramp01", "apbs_map01", [-5, 0, 5]) | |||
cmd.set("surface_ramp_above_mode", 1, "prepared01") | |||
cmd.set("surface_color", "apbs_ramp01", "prepared01") | |||
cmd.show("surface", "prepared01") | |||
</syntaxhighlight> | |||
== See Also == | == See Also == | ||
Latest revision as of 03:07, 18 May 2023
The APBS Electrostatics Plugin integrates the APBS software package into PyMOL. Its primary purpose is electrostatic surface visualization. It superseedes the APBS Tool2.1 plugin.
New in Incentive PyMOL 2.0
Usage
A molecule must already be loaded into PyMOL.
For regular protein and DNA molecules, the plugin should be a one-click solution (just click "Run").
For RNA, the residue names might need adjustment, see troubleshooting RNA.
Non-protein/DNA/RNA molecules or modified residues need special preparation, see #Prepare Molecule.
Procedure
The default procedure involves three steps:
- preparing the molecule by assigning partial charges and adding hydrogens and other missing atoms
- calculating the electrostatic map
- visualization of the charged molecular surface
Each step can be run individually by unchecking the checkboxes on the other steps.
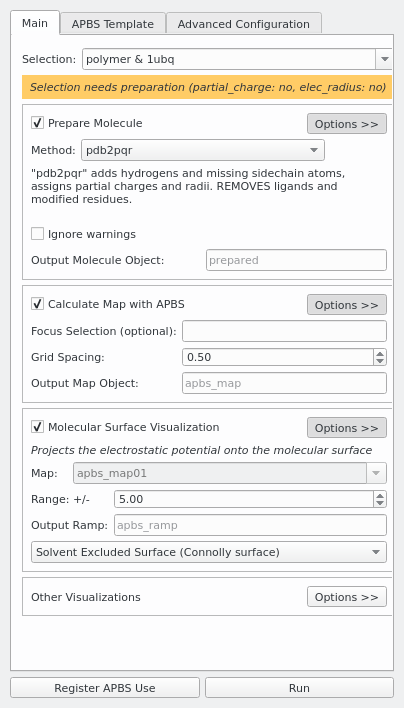
Prepare Molecule
APBS requires partial charges and atom radii. Since PDB files don't provide this information, they have to be preprocessed first.
pdb2pqr is the default preparation method, it is limited to protein and nucleic acid, unless the user provides his own forcefield parameters.
The prepwizard method is available if the user has the Schrodinger Suite installed. It can handle a variety of molecules.
You can also use any third-party tool that assigns partial charges and load the result into PyMOL. Supported file formats include PQR, MOL2, and MAE. In that case, uncheck "Prepare Molecule", or use "Method: use vdw" if you only have charges but no radii. Tools to consider are for example Open Babel or the PDB2PQR web server.
Calculate Map with APBS
- Focus Selection: If you only care about e.g. a binding site, you can specify a PyMOL selection here to speed up the calculation.
- Grid spacing (Angstrom): Increase to speed up the calculation and to reduce memory usage. Decrease to get a higher map resolution.
Molecular Surface Visualization
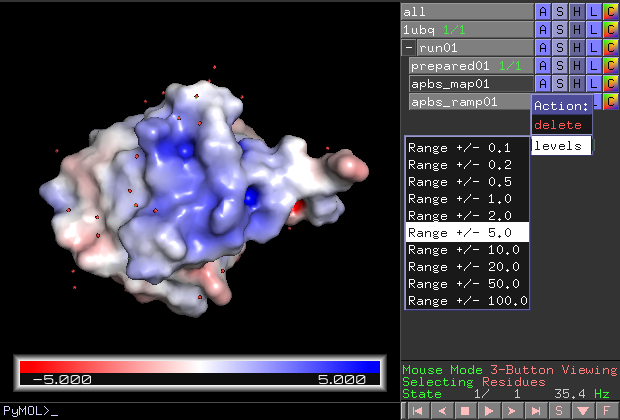
The most common visualization is to show the solvent excluded surface. PyMOL uses the potential at the solvent accessible surface for coloring in that case.
The range (color intensity) can also be changed later once the result is in PyMOL. Click "run01 > apbs_ramp01 > A > levels > ..." in the object menu panel.
Troubleshooting
RNA
For pdb2pqr, RNA resdiue names must be RA, RC, RG, and RU.
alter polymer & resn A+C+G+U, resn = "R" + resn
Incomplete Residues
Electrostatics should be analyzed on a complete molecule without missing sidechains, and ideally without missing loops. pdb2pqr will automatically model up to 10% of missing sidechain atoms. If your structure is incomplete, consider creating a homology model first using a tool of your choice (e.g. MODELLER).
Some truncated PDB files include a single backbone atom of the next residue, e.g. 2xwu chain B residue 954 atom N. pdb2pqr reports:
Error encountered: Too few atoms present to reconstruct or cap residue LEU B 954 in structure!
The easiest solution is to remove that atom:
remove /2xwu//B/954
APBS Template
Parameters like temperature, protein and solvent dielectric, or ion concentrations can be changed directly in the "APBS Template". Please read the APBS input file documentation.
Examples
RNA Molecule
fetch 1rna, async=0 alter polymer & resn A+C+G+U, resn = "R" + resn
- Open plugin
- click "Run"
Virus Assembly
set assembly, 1 fetch 3j7l, async=0 split_states 3j7l delete 3j7l
- Open plugin
- Selection: 3j7l_*
- Calculate Map with APBS > Options >> Grid Spacing: 2.0
- click "Run" (takes about 20 minutes!)
Python API
Disclaimer: The plugin code is not part of the official cmd API and could be subject to change.
Example with PDB 1ubq:
from pmg_tk.startup.apbs_gui.creating import pdb2pqr_cli
from pmg_tk.startup.apbs_gui.electrostatics import map_new_apbs
cmd.fetch("1ubq")
pdb2pqr_cli("prepared01", "1ubq", options=["--ff", "amber"])
map_new_apbs("apbs_map01", "prepared01")
cmd.ramp_new("apbs_ramp01", "apbs_map01", [-5, 0, 5])
cmd.set("surface_ramp_above_mode", 1, "prepared01")
cmd.set("surface_color", "apbs_ramp01", "prepared01")
cmd.show("surface", "prepared01")
See Also
- apbsplugin
- APBS
- ramp_new command