FindSurfaceCharge: Difference between revisions
Jump to navigation
Jump to search
TeddyWarner (talk | contribs) m (→Examples) |
TeddyWarner (talk | contribs) m (TeddyWarner moved page Findsurfaceresidueslistcharged to FindSurfaceResiduesListCharged: Correct capitalization) |
(No difference)
| |
Revision as of 12:00, 10 March 2023
| Type | Python Module |
|---|---|
| Download | findSurfaceResiduesListCharged.py |
| Author(s) | Teddy Warner |
| License | BSD-2-Clause |
| This code has been put under version control in the project Pymol-script-repo | |
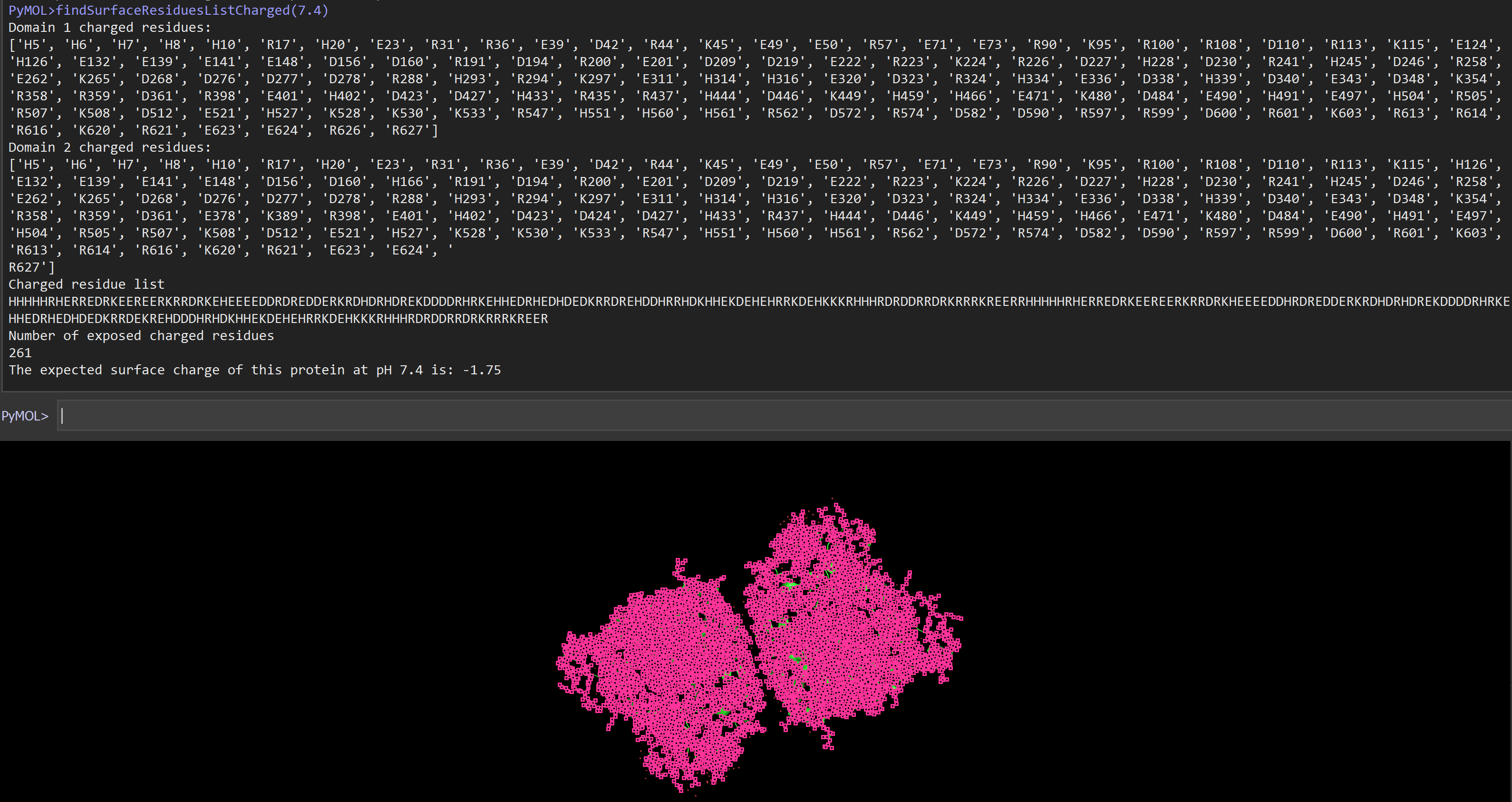
Based on the findSurfaceResidues script, the findSurfaceResiduesListCharged script will select and color surface residues and atoms on an object or selection. The function will also identify and output a list of all charged residues on the surface of a selection. This additionally calculates the ionization state of a protein at a given pH.
Each time, the script will create two new selections called, exposed_res_XYZ and exposed_atm_XYZ where XYZ is some random number. This is done so that no other selections/objects are overwritten.
Usage
findSurfaceResiduesListCharged [pH=7.0 [, selection=all [, cutoff=2.5 [, doShow=0 ]]]
Arguments
- pH = float: The pH to calculate the surface charge at {default: 7.0}
- selection = str: The object or selection for which to find exposed residues {default: all}
- cutoff = float: The cutoff in square Angstroms that defines exposed or not. Those atoms with > cutoff Ang^2 exposed will be considered exposed {default: 2.5 Ang^2}
- doShow = 0/1: Change the visualization to highlight the exposed residues vs interior {default: 0}
Examples
run findSurfaceResiduesListCharged.py
fetch 4FIX, async=0
findSurfaceResiduesListCharged
# see how pH changes the protein surface charge:
findSurfaceResiduesListCharged[7.2]
The expected surface charge of this protein at pH 7.2 is: -0.42
findSurfaceResiduesListCharged[10.0]
The expected surface charge of this protein at pH 10.0 is: -13.17
findSurfaceResiduesListCharged[5.0]
The expected surface charge of this protein at pH 5.0 is: +53.02