Fnab: Difference between revisions
Jump to navigation
Jump to search
(Creating Fnab page) |
No edit summary |
||
| Line 1: | Line 1: | ||
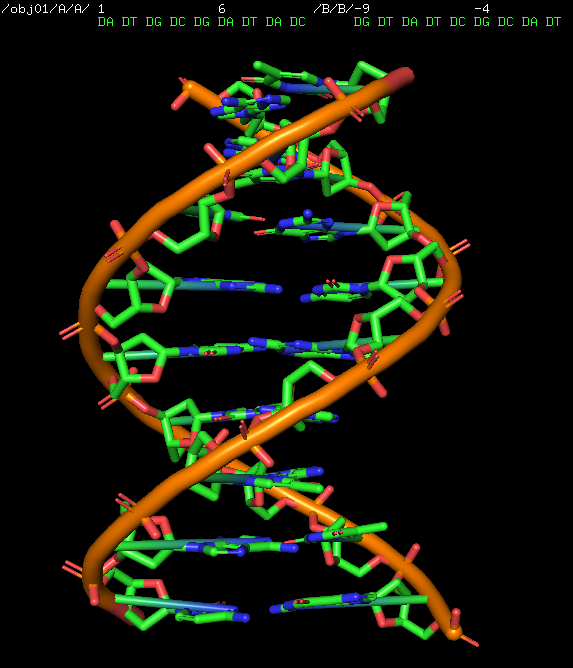
'''fnab''' builds nucleic acid entities from sequence. The sequence must be specified in one-letter code. Only one fragment can be created at a type (no spaces allowed unlike the 'fab' command). | '''fnab''' builds nucleic acid entities from sequence. The sequence must be specified in one-letter code. Only one fragment can be created at a type (no spaces allowed unlike the 'fab' command). | ||
Similar functionality is also provided by the graphical [[Builder]] in " | Similar functionality is also provided by the graphical [[Builder]] in "Nucleic Acid" mode. | ||
''New in PyMOL version 2.2'' | ''New in PyMOL version 2.2'' | ||
Revision as of 16:06, 18 December 2018
fnab builds nucleic acid entities from sequence. The sequence must be specified in one-letter code. Only one fragment can be created at a type (no spaces allowed unlike the 'fab' command).
Similar functionality is also provided by the graphical Builder in "Nucleic Acid" mode.
New in PyMOL version 2.2
USAGE
fnab input [, name [, type [, form [, dbl_helix ]]]]
ARGUMENTS
- input = string: Nucleic Acid sequence in one-letter code
- name = string: name of object to create or modify {default: obj??}
- mode = string: "DNA" or "RNA" {default: "DNA"}
- form = string: "A" or "B" {default: "B"}
- dbl_helix = bool (0/1): flag for using double helix in DNA
EXAMPLE
The two examples produce the same result:
# Create default (B-DNA) chain
fnab ATGCGATAC
# Create B-DNA chain with specific name
fnab ATGCGATAC, name=myDNA, mode=DNA, form=B, dbl_helix=1
# Create RNA chain
fnab AAUUUUCCG, mode=RNA