GROMACS Plugin: Difference between revisions
(→Gentoo) |
(Update) |
||
| Line 64: | Line 64: | ||
* GROMACS | * GROMACS | ||
* ProDy (optional) | * ProDy (optional) | ||
In order to take advantage of latest features you will need to have [http://www.csb.pitt.edu/prody/ ProDy] library installed. | * PLUMED (optional, compiled into GROMACS) | ||
In order to take advantage of latest features you will need to have [http://www.csb.pitt.edu/prody/ ProDy] library installed and [http://www.plumed.org/ PLUMED] compiled into GROMACS. | |||
== References == | == References == | ||
[http://pubs.acs.org/doi/abs/10.1021/ci400071x Molecular Dynamics Simulation by GROMACS Using GUI Plugin for PyMOL Tomasz Makarewicz and Rajmund Kaźmierkiewicz Journal of Chemical Information and Modeling 2013 53 (5), 1229-1234] | [http://pubs.acs.org/doi/abs/10.1021/ci400071x Molecular Dynamics Simulation by GROMACS Using GUI Plugin for PyMOL Tomasz Makarewicz and Rajmund Kaźmierkiewicz Journal of Chemical Information and Modeling 2013 53 (5), 1229-1234] | ||
[http://link.springer.com/article/10.1007/s00894-016-2982-4 Improvements in GROMACS plugin for PyMOL including implicit solvent simulations and displaying results of PCA analysis Tomasz Makarewicz, Rajmund Kaźmierkiewicz Journal of Molecular Modeling May 2016, 22:109] | |||
== License == | == License == | ||
Revision as of 07:57, 4 May 2016
| Type | PyMOL Plugin |
|---|---|
| Download | https://github.com/tomaszmakarewicz/Dynamics/releases |
| Author(s) | Laboratory of Biomolecular Systems Simulations |
| License | GPLv3 |
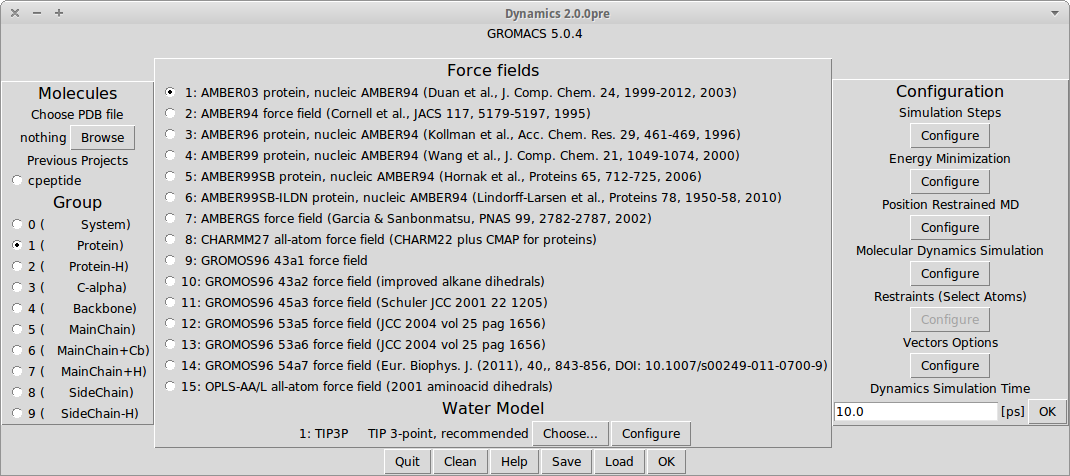
Dynamics PyMOL Plugin is plugin for PyMOL, which add molecular dynamics simulation feature. It is meant to be easy to use. The plugin uses GROMACS tools as a back-end. Project is developed as an open source and as such create full open source stack together with PyMOL and GROMACS. Because GROMACS can compile easily only on Unix like systems, the plugin also keep compatibility only with those. No Windows port is expected.
Website
https://github.com/tomaszmakarewicz/Dynamics
Features
- Easy to use GUI, to take advantage of complex software GROMACS.
- Work directly on molecules loaded to PyMOL.
- Display results of calculations directly in PyMOL.
- Set restraints, choose water models, force fields and many more.
- Save your work and finish calculations later or on the other machine.
- Do your work free of charge and without any restrictions. Feel free to modify any component of the stack.
- Minimum dependencies. Plugin use the same graphic libraries as PyMOL, so working PyMOL and GROMACS installations are enough to make plugin work.
Note that exact instruction of how to use program is in the software manual, which is available together with the plugin.
Installation
There are at least few ways to install the plugin. In this section we will describe the most common. Please keep in mind, that this plugin is develop for Unix platform - it will not work on MS Windows.
Ubuntu
The easiest way to install the plugin is to use PPA repositories:
sudo add-apt-repository ppa:tomaszm/dynamics sudo apt-get update sudo apt-get install pymol-plugin-dynamics
All required libraries will be downloaded as dependencies. Supported versions of Ubuntu are latest stable and latest LTS.
Gentoo
The plugin is also present in Gentoo ebuild system. To make the installation use (as root):
emerge --ask sci-chemistry/pymol-plugins-dynamics
Other GNU/Linux and Unix
The second way for all other platforms or Ubuntu as well, is to download latest release of the plugin directly from its GitHub webpage:
https://github.com/tomaszmakarewicz/Dynamics/releases
Then run PyMOL as a root. On the top Menu choose Plugin->Manage Plugins->Install... Then choose pymol_plugin_dynamics.py file. After installation restart PyMOL with normal user privilege.
Required dependencies:
- PyMOL
- GROMACS
- ProDy (optional)
In order to take advantage of latest features you will need to have ProDy library installed.
Latest Snapshots
You can download latest master snapshot by command:
git clone git://github.com/tomaszmakarewicz/Dynamics.git
Then run PyMOL as a root. On the top Menu choose Plugin->Manage Plugins->Install... Then choose pymol_plugin_dynamics.py file. After installation restart PyMOL with normal user privilege.
Required dependencies:
- PyMOL
- GROMACS
- ProDy (optional)
- PLUMED (optional, compiled into GROMACS)
In order to take advantage of latest features you will need to have ProDy library installed and PLUMED compiled into GROMACS.
References
Molecular Dynamics Simulation by GROMACS Using GUI Plugin for PyMOL Tomasz Makarewicz and Rajmund Kaźmierkiewicz Journal of Chemical Information and Modeling 2013 53 (5), 1229-1234 Improvements in GROMACS plugin for PyMOL including implicit solvent simulations and displaying results of PCA analysis Tomasz Makarewicz, Rajmund Kaźmierkiewicz Journal of Molecular Modeling May 2016, 22:109
License
This software (including its Debian packaging) is available to you under the terms of the GPL-3, see "/usr/share/common-licenses/GPL-3". Software is created and maintained by Laboratory of Biomolecular Systems Simulation at University of Gdansk.
Contributors:
- Tomasz Makarewicz (btchtm [at] ug [dot] edu [dot] pl)