Valence: Difference between revisions
No edit summary |
(added information to the valence command and distinguished it from the settings along with minor edits) |
||
| Line 1: | Line 1: | ||
== Overview == | == Overview == | ||
Turning on '''valence''' will enable the display of double bonds.<br> | Turning on the '''valence''' setting will enable the display of double bonds.<br> | ||
Toggling '''valence_mode''' alters the positioning of double bonds (for representation as [[Lines]])<br> | Toggling '''valence_mode''' alters the positioning of double bonds (for representation as [[Lines]])<br> | ||
'''valence_size''' alters the distance of double bonds.<br> | '''valence_size''' alters the distance of double bonds.<br> | ||
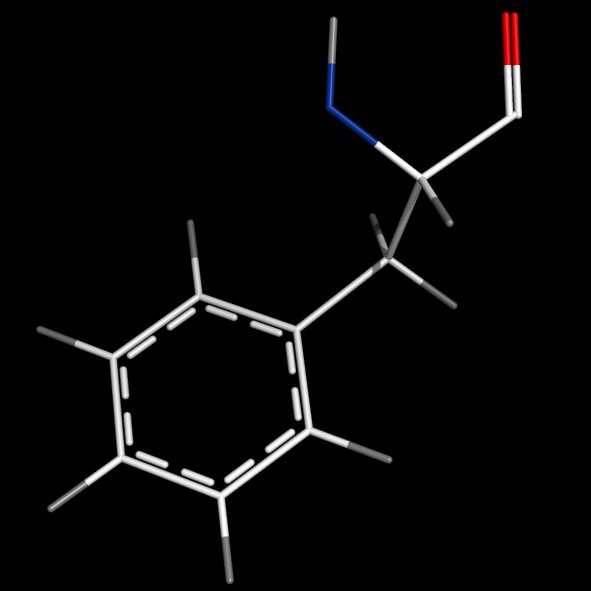
Note that bonds can be edited to be delocalized using [[Unbond]] and [[Bond]].<br> | Note that bonds can be edited to be delocalized using [[Unbond]] and [[Bond]].<br> | ||
There is also a command called '''valence'''.<br> | |||
== Examples == | == Examples for the settings: valence and valence_mode == | ||
<gallery heights="200px" widths="200px" align="left" perrow="4"> | <gallery heights="200px" widths="200px" align="left" perrow="4"> | ||
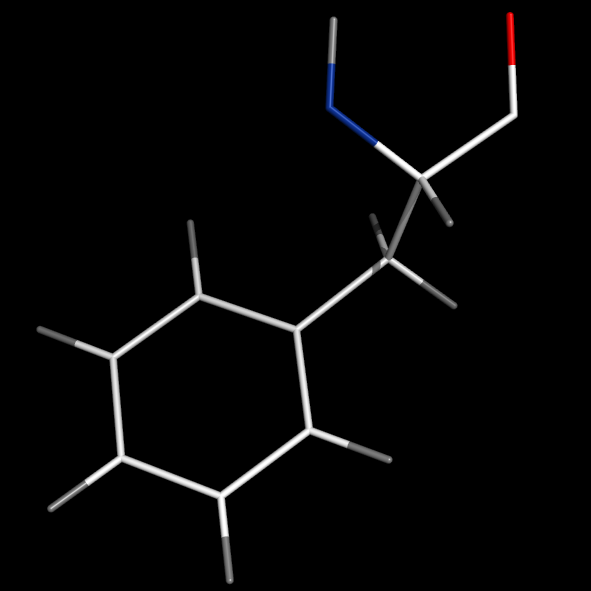
Image:PHE_valence_0.png|set valence, 0<br>#(no double bonds) | Image:PHE_valence_0.png|set valence, 0<br>#(no double bonds) | ||
| Line 22: | Line 23: | ||
||[[File:valence_size_mode1.gif]] || [[File:valence_size_mode0.gif]] | ||[[File:valence_size_mode1.gif]] || [[File:valence_size_mode0.gif]] | ||
|} | |} | ||
=== Syntax === | |||
== Syntax == | |||
<source lang="python"> | <source lang="python"> | ||
set valence, 0 # off | set valence, 0 # off | ||
| Line 32: | Line 31: | ||
set valence_mode, 1 # inside | set valence_mode, 1 # inside | ||
set valence_size, 0.1 # default: 0.06 # range 0 - ~0.5 | set valence_size, 0.1 # default: 0.06 # range 0 - ~0.5 | ||
</source> | |||
== The valence command == | |||
The '''valence''' command automatically formats existing bonds and can even guess the bonds for standard amino acids.<br> | |||
<source lang="python"> | |||
#USAGE: | |||
valence order, selection1 [, selection2 [, source [, target_state [, source_state [, reset [, quiet ]]]]]] | |||
order can be either: 1, 2, 3, 4, aromatic, copy, guess | |||
# Editing bonds | #make PyMOL guess/autoformat bonds in proteins | ||
valence guess, all | |||
</source> | |||
== Editing bonds == | |||
<source lang="python"> | |||
# In editing mode: select the bond using Ctrl-right-click, then enter: | # In editing mode: select the bond using Ctrl-right-click, then enter: | ||
unbond pk1,pk2 | unbond pk1,pk2 | ||
| Line 43: | Line 57: | ||
==Automatic editing of bonds== | ==Automatic editing of bonds== | ||
[[Format_bonds]] is a script that automatically formats valence in amino acids. | Try using the '''valence''' command first.<br> | ||
Secondly, [[Format_bonds]] is a script that automatically formats valence in all amino acids and has additional options. | |||
Revision as of 05:01, 22 April 2015
Overview
Turning on the valence setting will enable the display of double bonds.
Toggling valence_mode alters the positioning of double bonds (for representation as Lines)
valence_size alters the distance of double bonds.
Note that bonds can be edited to be delocalized using Unbond and Bond.
There is also a command called valence.
Examples for the settings: valence and valence_mode
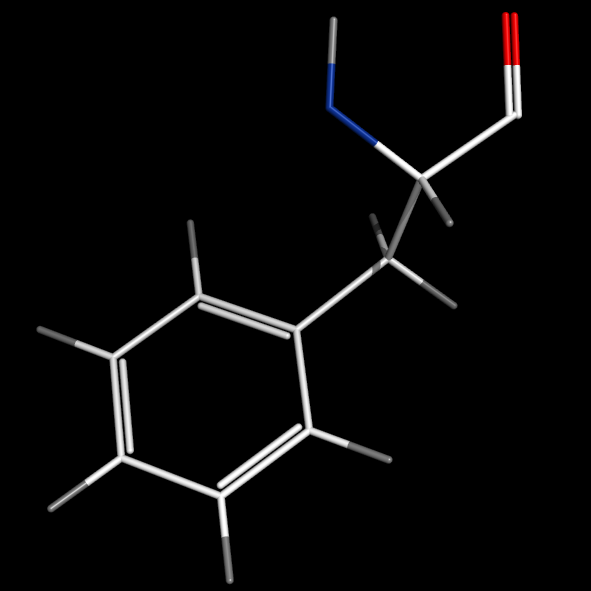
set valence, 1
#delocalized bonds
#(edited: see Bond)
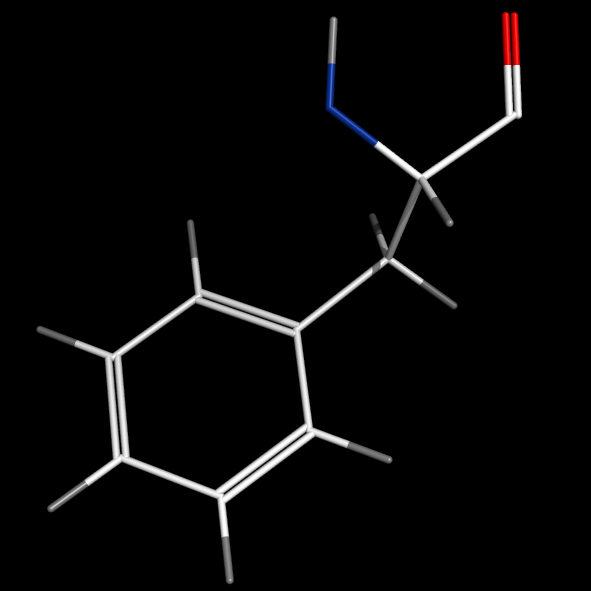
valence_size alters the distance of double bonds, but behaves slightly different depending on valence_mode
| valence_size with valence_mode 1 inside |
valence_size with valence_mode 0 centered |
|---|---|
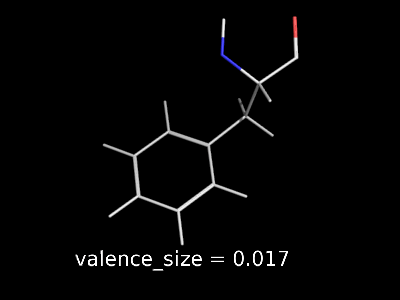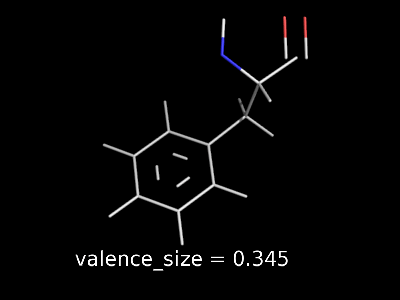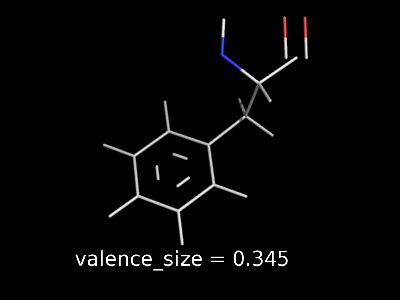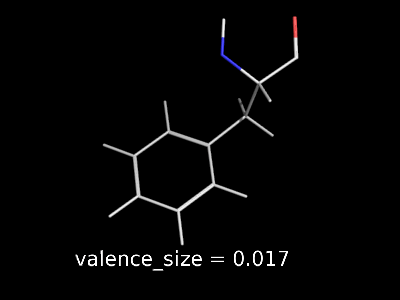 |
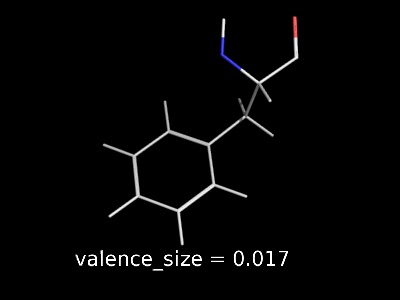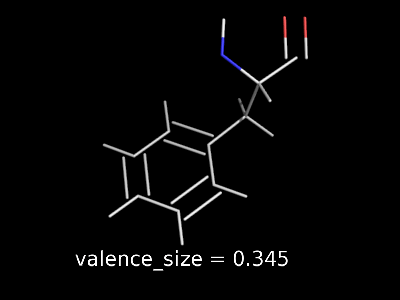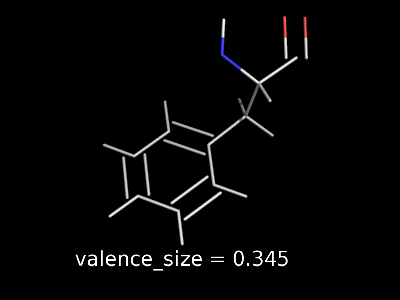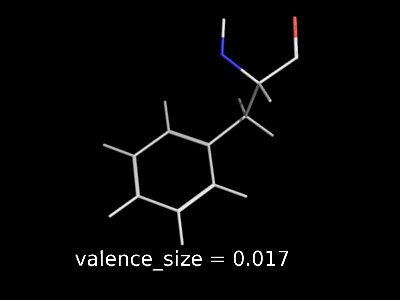
|
Syntax
set valence, 0 # off
set valence, 1 # on
set valence_mode, 0 # centered
set valence_mode, 1 # inside
set valence_size, 0.1 # default: 0.06 # range 0 - ~0.5
The valence command
The valence command automatically formats existing bonds and can even guess the bonds for standard amino acids.
#USAGE:
valence order, selection1 [, selection2 [, source [, target_state [, source_state [, reset [, quiet ]]]]]]
order can be either: 1, 2, 3, 4, aromatic, copy, guess
#make PyMOL guess/autoformat bonds in proteins
valence guess, all
Editing bonds
# In editing mode: select the bond using Ctrl-right-click, then enter:
unbond pk1,pk2
bond pk1,pk2,4
# 1: single bond, 2: double bond, 3:triple bond, 4:delocalized
Automatic editing of bonds
Try using the valence command first.
Secondly, Format_bonds is a script that automatically formats valence in all amino acids and has additional options.