Cartoon: Difference between revisions
| Line 124: | Line 124: | ||
</source> | </source> | ||
=== Other Nucleic Acids Settings === | === Other Nucleic Acids & Cartoon Settings === | ||
Here are some things to try: | Here are some things to try: | ||
<source lang="python"> | <source lang="python"> | ||
Revision as of 13:45, 8 November 2005
Cartoon Command
DESCRIPTION
"cartoon" changes the default cartoon for a set of atoms.
USAGE
cartoon type, (selection) type = skip | automatic | loop | rectangle | oval | tube | arrow | dumbbell
PYMOL API
cmd.cartoon(string type, string selection )
EXAMPLES
cartoon rectangle,(chain A) cartoon skip,(resi 145:156)
NOTES
the "automatic" mode utilizes ribbons according to the information in the PDB HELIX and SHEET records.
Adjusting width of cartoon
Try varying the following.
For β-strands:
cartoon_rect_length
cartoon_rect_width
For α-helices:
cartoon_oval_length
cartoon_oval_width
For loops:
cartoon_loop_radius
For "fancy" α-helices:
cartoon_dumbell_length
cartoon_dumbell_width
cartoon_dumbell_radius (radius of cylinder at edge of helix ribbon)
In each case "length" refers to what some might call the width and "width" refers to what some might call the thickness.
Forcing Exact Boundaries in Coloring Secondary Structures
To force PyMol to respect secondary structural elements color-wise (PyMol smooths out colors near color chagnes for a prettier image) use the following
PyMol command:
set cartoon_discrete_colors, on
Sausage Representation
The familiar sausage representation in PyMol is called, "putty". To enable the putty/sausage view simply do,
show cartoon cartoon putty unset cartoon_smooth_loops unset cartoon_flat_sheets
As of v 0.98 or so, there's a Putty option. Use this.
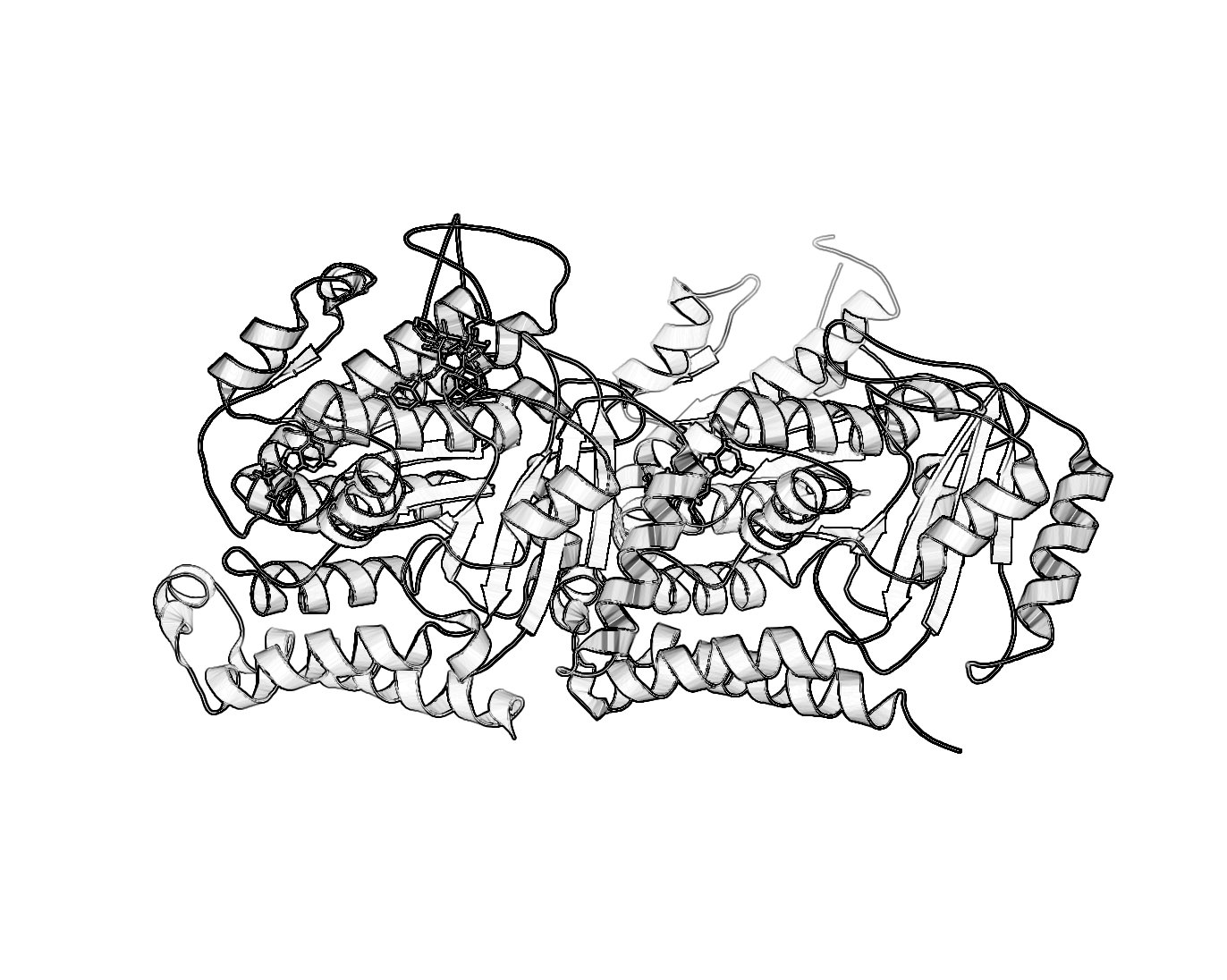
Black and White Representation
For those who want a nifty black and white representation of their protein try the following:
- Ray trace your protein of choice in a cartoon representation use a BLACK background
- Save the image
- Load the image in GIMP.
- Desaturate or Grayscale the image.
- Run the filter: Filter->Edge-Detect->Edge.
- Select: Layers->Color->Invert.
- Different methods of edge detection will give you different results. In the last example, I used Laplace Edge-Detect, then painted an all white layer beneath the current layer to achieve the results.
I'm sure there are other ways to do this. If you want to include it in a publication make sure you ray traced it large enough. For that, see Creating Publication Quality Images.
CA (Alpha Carbon) Trace
If you have a structure with just a alpha carbon trace, you can get a cartoon by
set cartoon_trace,1
show cartoon
If your structure is more than just the CA backbone, the cartoon representation will look incorrect, so use it just with CA trace.
Various Transparency Levels
One can make different cartoon selections have different transparency values, in PyMol. The trick here is to use "create" instead of "select". Create makes new objects that can have independent settings.
load mol_obj.pdb
# transfer a piece of the molecule into a new object
create new_obj, chain A
remove mol_obj in new_obj
# adjust trasparency for the new object
set cartoon_transparency, 0.5, new_obj
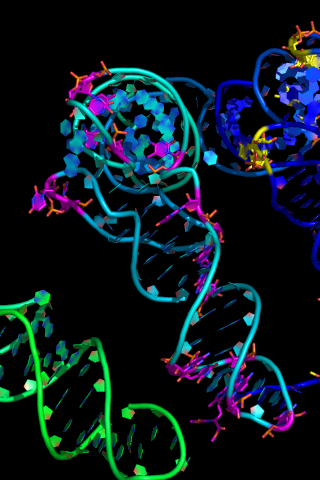
Nucleic Acid Representation
To show nucleic acids in a nicer format do:
set cartoon_ring_mode,1
show cartoon
Other Nucleic Acids & Cartoon Settings
Here are some things to try:
set cartoon_ring_mode, 1 # (or 2 or 3)
set cartoon_ring_finder, 1 # (or 2 or 3 or 4)
set nucleic_acid_mode, 0 # (or 1 or 2 or 3 or 4)
set cartoon_side_chain_helper
rebuild
set cartoon_ring_transparency, 0.5
set cartoon_ladder_mode, 0 # or 1
set cartoon_ladder_color, color-name
set cartoon_nucleic_acid_color, color-name
cartoon oval
set cartoon_oval_width, 0.8
cartoon rect
cartoon dumbbell
set cartoon_dumbbell_width, 0.4
set cartoon_dumbbell_radius, 0.4