Sidechaincenters: Difference between revisions
Appearance
figure added |
No edit summary |
||
| Line 135: | Line 135: | ||
</source> | </source> | ||
[[Category:Script_Library]] | [[Category:Script_Library]] | ||
[[Category:Structural_Biology_Scripts]] | [[Category:Structural_Biology_Scripts]] | ||
Revision as of 20:17, 13 December 2011
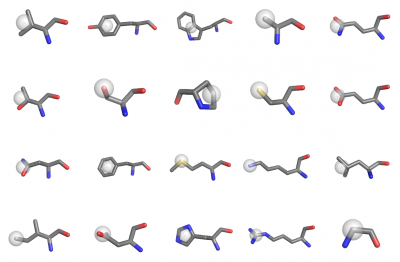
Pseudo single-atom representation of sidechains. Usefull for pair potential calculation for example.
Example
fetch 2x19
sidechaincenters scc, 2x19
The Script
'''
(c) 2010 Thomas Holder
'''
from pymol import cmd
from chempy import Atom, cpv, models
sidechaincenteratoms = {
'GLY': ('CA',),
'ALA': ('CB',),
'VAL': ('CG1', 'CG2'),
'ILE': ('CD1',),
'LEU': ('CD1', 'CD2'),
'SER': ('OG',),
'THR': ('OG1', 'CG2'),
'ASP': ('OD1', 'OD2'),
'ASN': ('OD1', 'ND2'),
'GLU': ('OE1', 'OE2'),
'GLN': ('OE1', 'NE2'),
'LYS': ('NZ',),
'ARG': ('NE', 'NH1', 'NH2'),
'CYS': ('SG',),
'MET': ('SD',),
'MSE': ('SE',),
'PHE': ('CG', 'CD1', 'CD2', 'CE1', 'CE2', 'CZ'),
'TYR': ('CG', 'CD1', 'CD2', 'CE1', 'CE2', 'CZ', 'OH'),
'TRP': ('CG', 'CD1', 'CD2', 'NE1', 'CE2', 'CE3', 'CZ2', 'CZ3'),
'HIS': ('CG', 'ND1', 'CD2', 'CE1', 'NE2'),
'PRO': ('CB', 'CG', 'CD'),
}
def sidechaincenters(object='scc', selection='all', name='PS1', method='bahar1996'):
'''
DESCRIPTION
Creates an object with sidechain representing pseudoatoms for each residue
in selection.
Sidechain interaction centers as defined by Bahar and Jernigan 1996
http://www.ncbi.nlm.nih.gov/pubmed/9080182
USAGE
sidechaincenters object [, selection]
ARGUMENTS
object = string: name of object to create
selection = string: atoms to consider {default: (all)}
name = string: atom name of pseudoatoms {default: PS1}
SEE ALSO
sidechaincentroids, pseudoatom
'''
atmap = dict()
if method == 'bahar1996':
modelAll = cmd.get_model('(%s) and resn %s' % (selection, '+'.join(sidechaincenteratoms)))
for at in modelAll.atom:
if at.name in sidechaincenteratoms[at.resn]:
atmap.setdefault((at.segi, at.chain, at.resn, at.resi), []).append(at)
elif method == 'centroid':
modelAll = cmd.get_model('(%s) and not (hydro or name C+N+O)' % selection)
for at in modelAll.atom:
atmap.setdefault((at.segi, at.chain, at.resn, at.resi), []).append(at)
else:
print 'Error: unknown method:', method
return
model = models.Indexed()
for centeratoms in atmap.itervalues():
center = cpv.get_null()
for at in centeratoms:
center = cpv.add(center, at.coord)
center = cpv.scale(center, 1./len(centeratoms))
atom = Atom()
atom.coord = center
atom.index = model.nAtom + 1
atom.name = name
for key in ['resn','chain','resi','resi_number','hetatm','ss','segi']:
atom.__dict__[key] = at.__dict__[key]
model.add_atom(atom)
model.update_index()
if object in cmd.get_object_list():
cmd.delete(object)
cmd.load_model(model, object)
return model
def sidechaincentroids(object='scc', selection='all', name='PS1'):
'''
DESCRIPTION
Sidechain centroids. Works like "sidechaincenters", but the
pseudoatom is the centroid of all atoms except hydrogens and backbone atoms
(N, C and O).
NOTE
If you want to exclude C-alpha atoms from sidechains, modify the selection
like in this example:
sidechaincentroids newobject, all and (not name CA or resn GLY)
SEE ALSO
sidechaincenters
'''
return sidechaincenters(object, selection, name, method='centroid')
cmd.extend('sidechaincenters', sidechaincenters)
cmd.extend('sidechaincentroids', sidechaincentroids)
cmd.auto_arg[1].update({
'sidechaincenters' : [ cmd.selection_sc , 'selection' , '' ],
'sidechaincentroids' : [ cmd.selection_sc , 'selection' , '' ],
})
# vi: ts=4:sw=4:smarttab:expandtab