Dssp stride: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
|||
| Line 2: | Line 2: | ||
{{Infobox script-repo | {{Infobox script-repo | ||
|type = Python Module | |type = Python Module | ||
|filename = | |filename = plugins/dssp_stride.py | ||
|author = [[User:Hongbo_zhu|Hongbo Zhu Cn]] | |author = [[User:Hongbo_zhu|Hongbo Zhu Cn]] | ||
|license = - | |license = - | ||
Revision as of 07:00, 11 December 2011
Introduction
| Type | Python Module |
|---|---|
| Download | plugins/dssp_stride.py |
| Author(s) | Hongbo Zhu Cn |
| License | - |
| This code has been put under version control in the project Pymol-script-repo | |
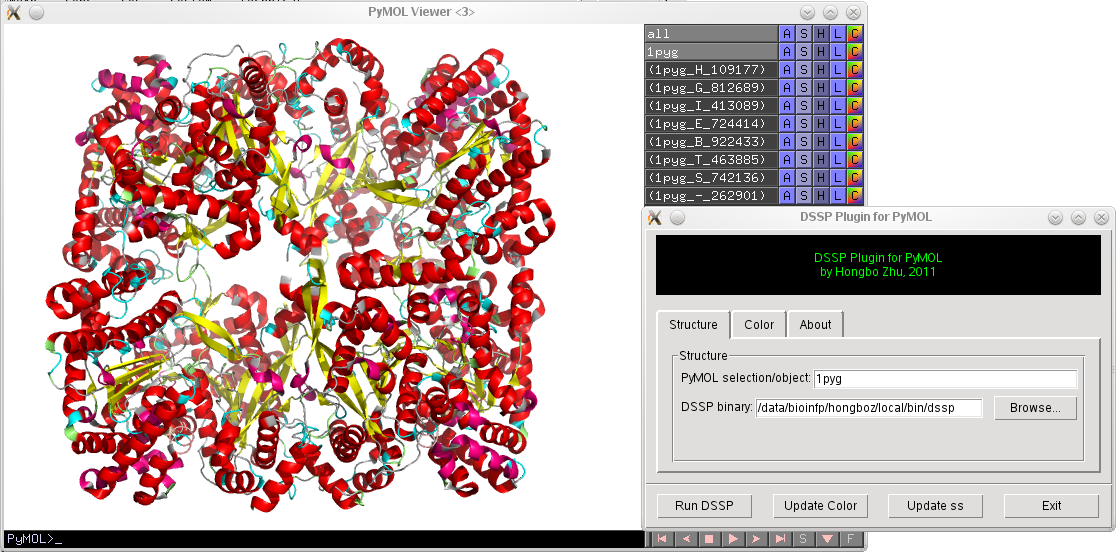
DSSP is a popular tool for assigning secondary structures for proteins. DSSP Plugin for PyMOL provides a graphical user interface for coloring proteins according to their secondary structures assigned by DSSP. DSSP Plugin can be obtained from: http://www.biotec.tu-dresden.de/~hongboz/dssp_pymol/dssp_pymol.html
Reference
http://www.biotec.tu-dresden.de/~hongboz/dssp_pymol/dssp_pymol.html