Color by conservation: Difference between revisions
Jump to navigation
Jump to search
mNo edit summary |
m (d'oh) |
||
| Line 71: | Line 71: | ||
c = float(1.0 + len(af)) / float(M) | c = float(1.0 + len(af)) / float(M) | ||
for y in af: | for y in af: | ||
_self.alter("%s and | _self.alter("%s and index %s" % (y[0], y[1]), "b=c", space={'c':c}) | ||
if as_putty!=0: | if as_putty!=0: | ||
Revision as of 11:50, 8 December 2011
Overview
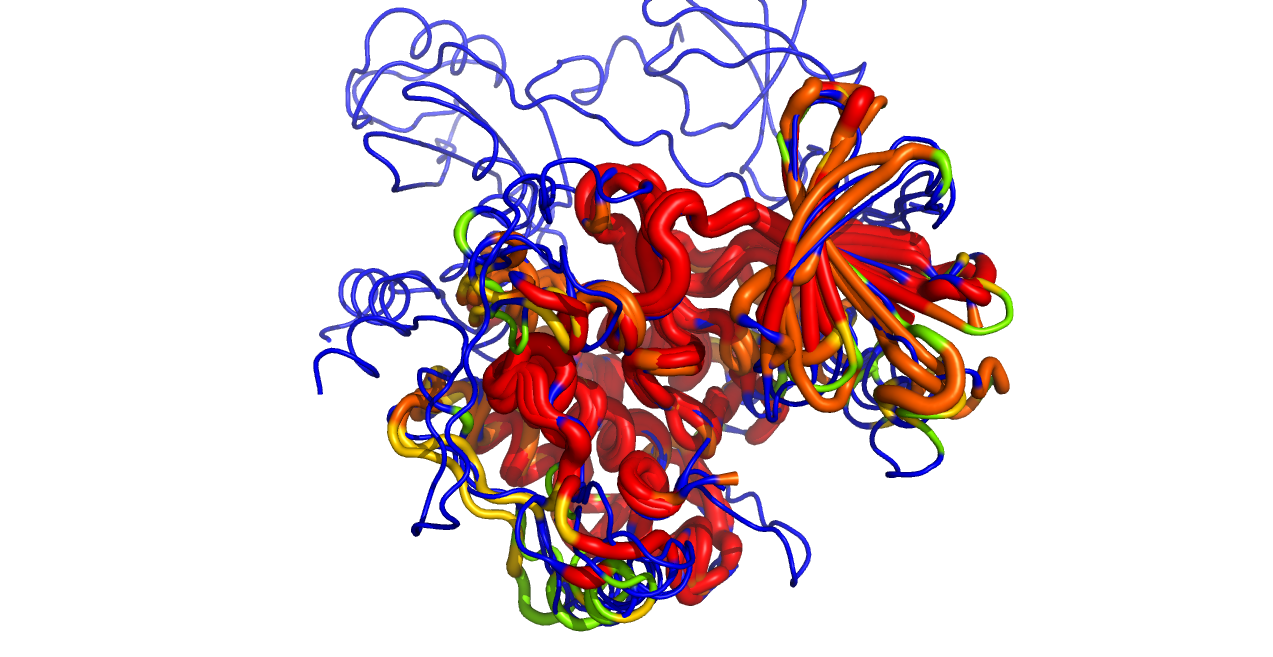
This script reads an alignment object and colors the protein objects in the alignment by the sequence conservation found in the alignment.
Example Usage
# run the script
run show_conserved.py
# turn on the sequence viewer
set seq_view
# get some kinases
fetch 1opk 3dtc 3p86 2eva 3efw, async=0
# align them into the "aln" object
for x in cmd.get_names(): cmd.align(x, "3efw and c. A", object="aln")
# color
color_by_conservation("aln", as_putty=1)
The Code
def color_by_conservation(aln, names=(), color="rainbow", as_putty=0, _self=cmd):
"""
PARAMETERS
aln
(string) the alignment object name
names
(list) a list of object names that are in the alignment;
if (), then PyMOL will attempt to glean the names
from the alignment object
color
(string) valid PyMOL spectrum name
as_putty
(0 or 1) if 0 display is not changed, else participating objects are shown
as cartoon putty, colored by the 'color' field
"""
# PyMOL doesn't yet know about object:alignment
# but we need to check that this exists or we might crash
if _self.get_type(aln)!="object:":
print "Error: Bad or incorrectly specified alignment object."
return None
r = cmd.get_raw_alignment(aln)
if names==():
known_objs = []
map(known_objs.extend, map(lambda x: map(lambda y: y[0], x), r))
known_objs=set(known_objs)
# highest number of matches seen
M = max(map(len,r)) + 1
else:
known_objs = set(names)
M = len(known_objs) + 1
for obj in known_objs:
_self.alter(obj, "b=0.0")
for af in r:
c = float(1.0 + len(af)) / float(M)
for y in af:
_self.alter("%s and index %s" % (y[0], y[1]), "b=c", space={'c':c})
if as_putty!=0:
for obj in known_objs:
_self.show_as("cartoon", "%s" % obj)
_self.cartoon("putty", "%s" % obj)
_self.spectrum('b', color, obj)
_self.sort()
_self.rebuild()
return None