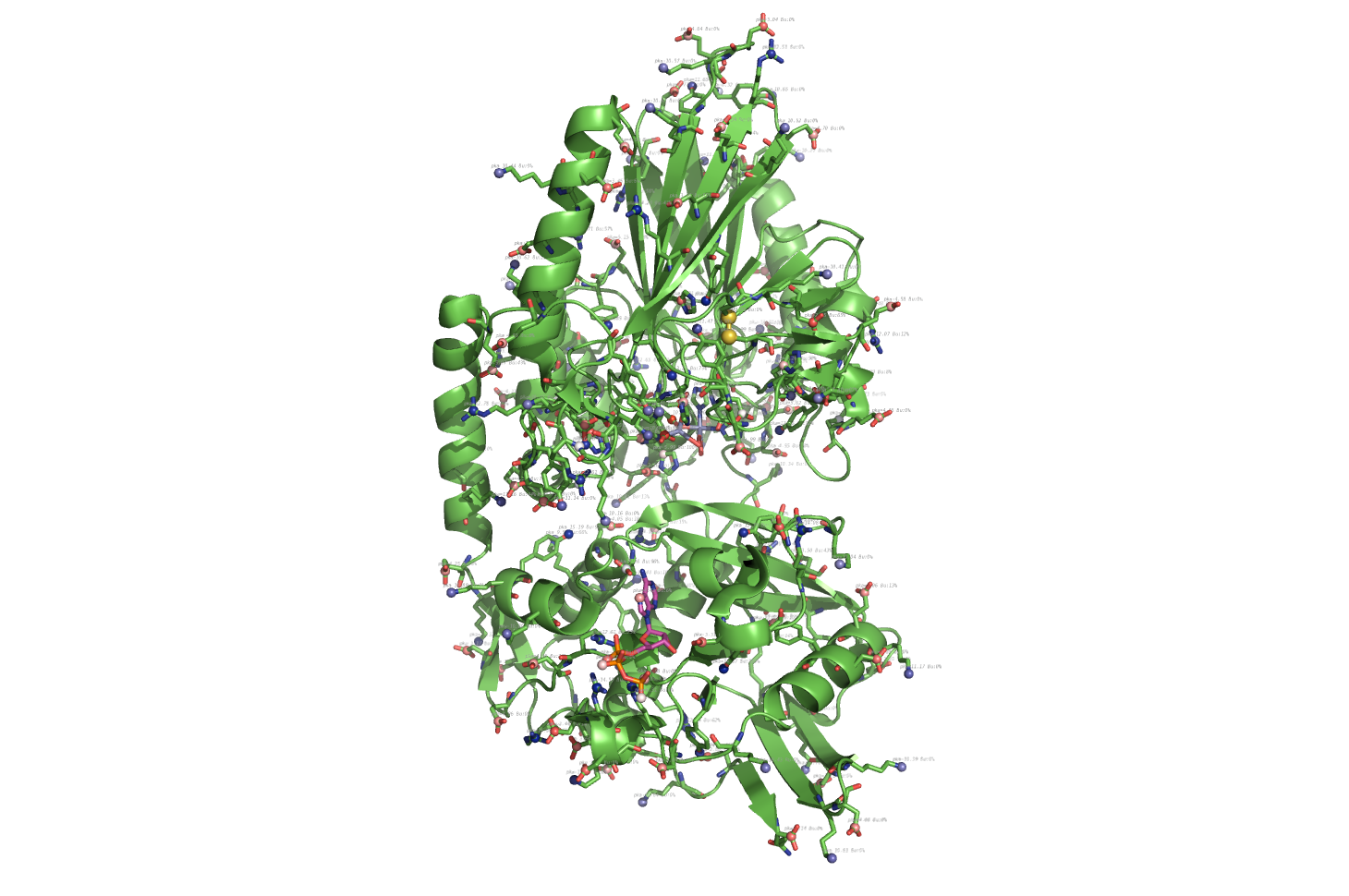
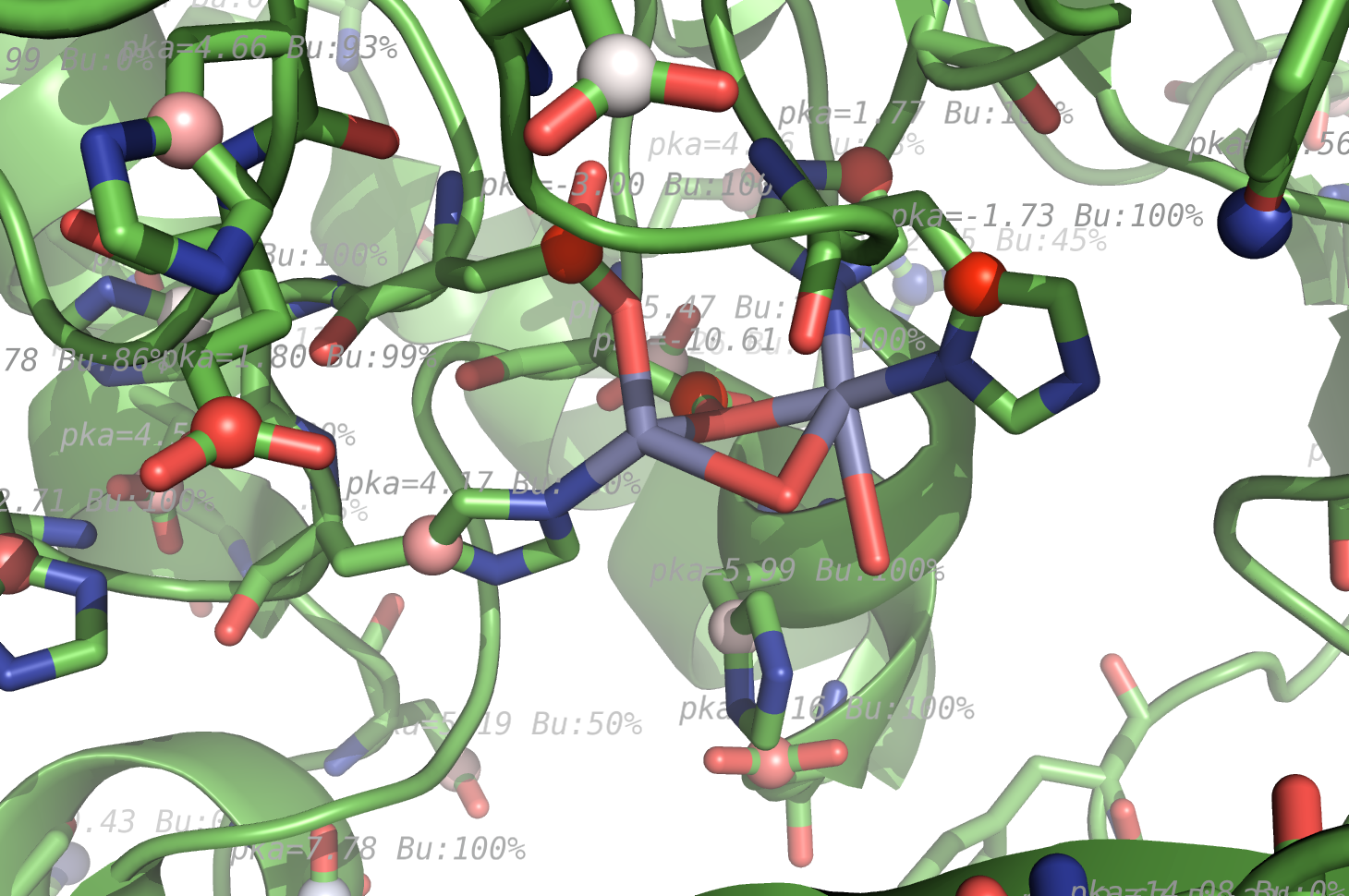
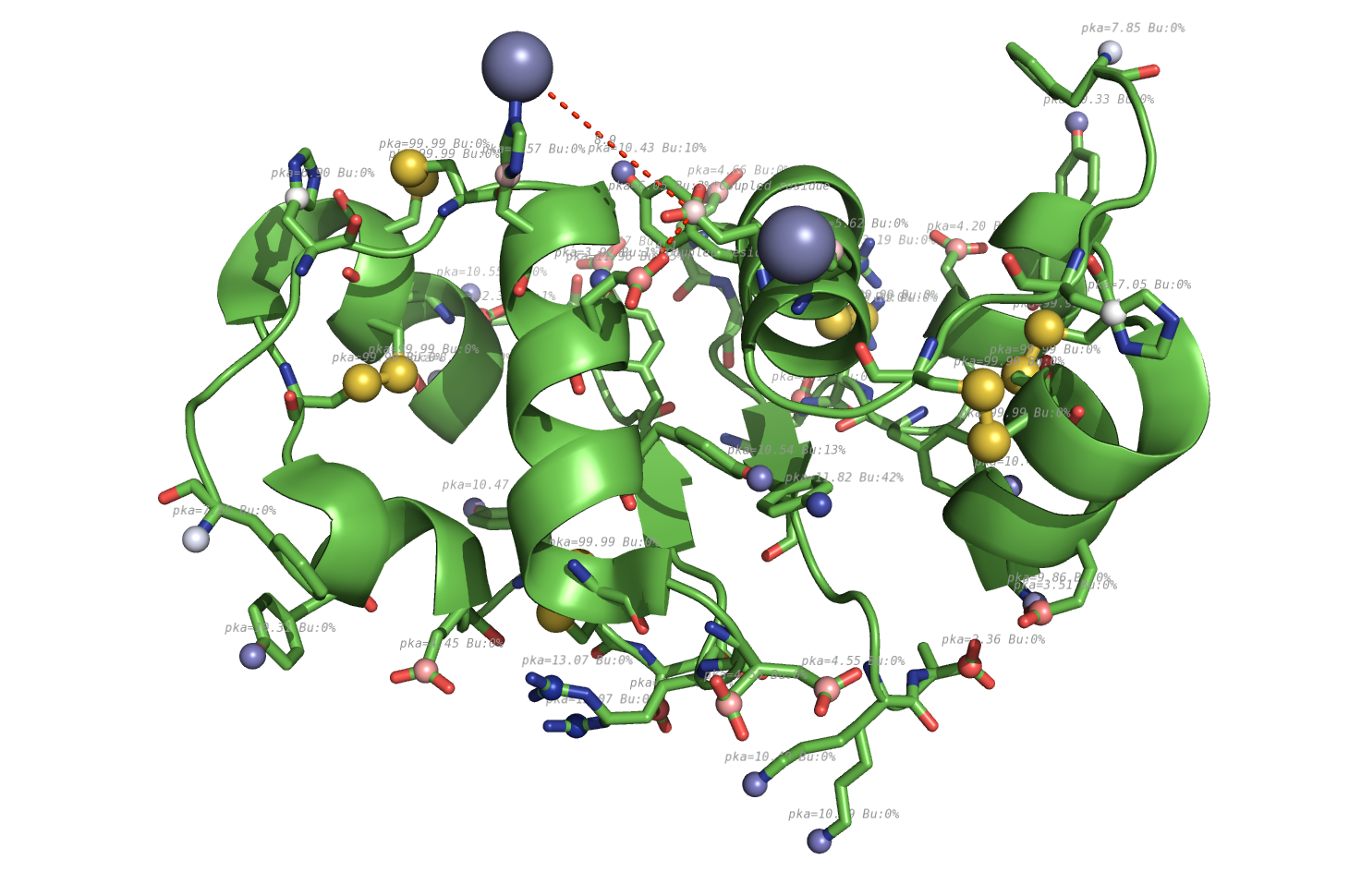
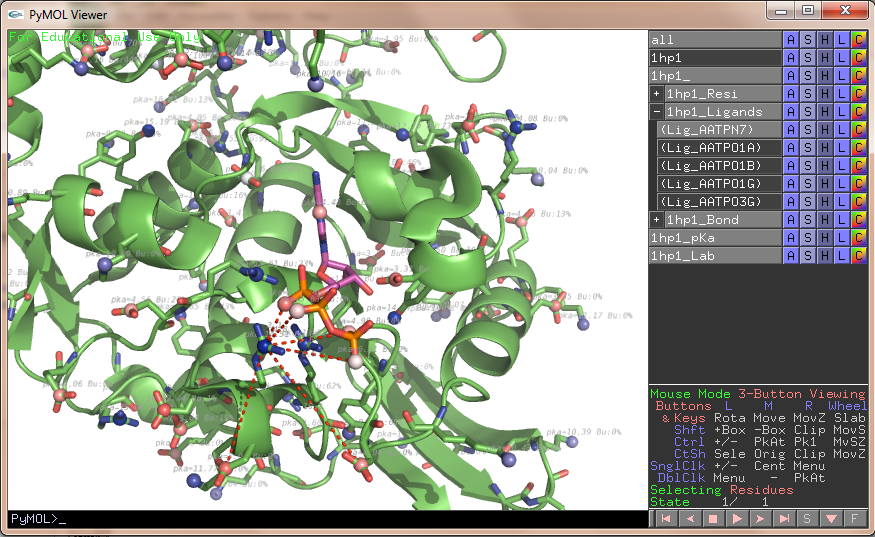
|
|
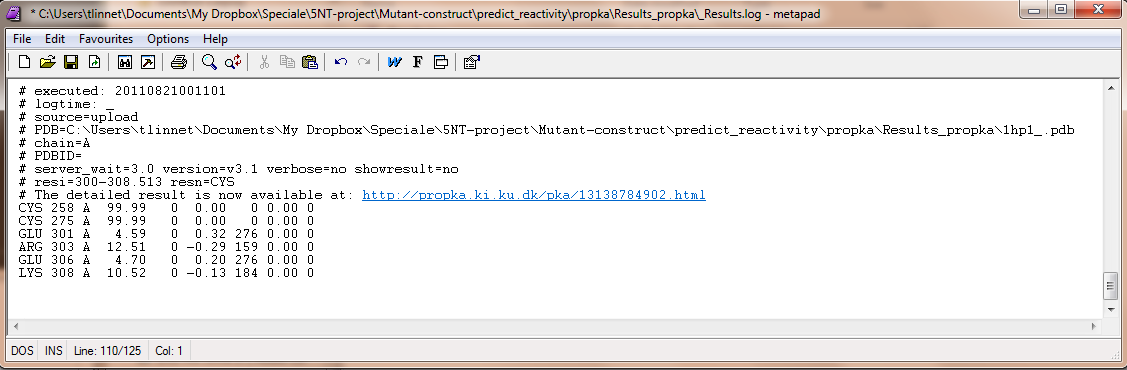
| Line 249: |
Line 249: |
|
| |
|
| == Python Code == | | == Python Code == |
| The code can be downloaded fast from here http://tinyurl.com/pymolpropka. <br />
| | This code has been put under version control. In the project, [http://www.pymolwiki.org/index.php/Git_intro Pymol-script-repo]. |
| # wget http://tinyurl.com/pymolpropka
| |
| # mv pymolpropka propka.py
| |
|
| |
|
| <syntaxhighlight lang="python">
| | For a color coded view: |
| #-------------------------------------------------------------------------------
| | https://github.com/Pymol-Scripts/Pymol-script-repo/blob/master/propka.py |
| # Name: propka for pymol
| | See the raw code or download manually, by right clicking the following link here -> Save as: propka.py |
| # Purpose: To fetch and display the pka values for protein of intetest
| | https://raw.github.com/Pymol-Scripts/Pymol-script-repo/master/propka.py |
| #
| |
| # Author: Troels E. Linnet
| |
| #
| |
| # Created: 14/08/2011
| |
| # Copyright: (c) Troels E. Linnet 2011
| |
| # Contact: tlinnet snabela gmail dot com
| |
| # Licence: Free for all
| |
| #
| |
| # Download: http://tinyurl.com/pymolpropka
| |
| #
| |
| #-------------------------------------------------------------------------------
| |
| """
| |
| The PROPKA method is developed by the
| |
| Jensen Research Group
| |
| Department of Chemistry
| |
| University of Copenhagen
| |
| | |
| Please cite these references in publications:
| |
| Hui Li, Andrew D. Robertson, and Jan H. Jensen
| |
| "Very Fast Empirical Prediction and Interpretation of Protein pKa Values"
| |
| Proteins, 2005, 61, 704-721.
| |
| | |
| Delphine C. Bas, David M. Rogers, and Jan H. Jensen
| |
| "Very Fast Prediction and Rationalization of pKa Values for Protein-Ligand Complexes"
| |
| Proteins, 2008, 73, 765-783.
| |
| | |
| Mats H.M. Olsson, Chresten R. Soendergard, Michal Rostkowski, and Jan H. Jensen
| |
| "PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa predictions"
| |
| Journal of Chemical Theory and Computation, 2011 7 (2), 525-537
| |
| | |
| Chresten R. Soendergaard, Mats H.M. Olsson, Michaz Rostkowski, and Jan H. Jensen
| |
| "Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of pKa Values"
| |
| Journal of Chemical Theory and Computation, 2011 in press
| |
| """
| |
| #-------------------------------------------------------------------------------
| |
| # The script needs mechanize to run.
| |
| # On windows, it is not easy to make additional modules available for pymol. So put in into your working folder.
| |
| #1)The easy manual way:
| |
| #a)Go to: http://wwwsearch.sourceforge.net/mechanize/download.html
| |
| #b)Download mechanize-0.2.5.zip. http://pypi.python.org/packages/source/m/mechanize/mechanize-0.2.5.zip
| |
| #c)Extract to .\mechanize-0.2.5 then move the in-side folder "mechanize" to your folder with propka.py. The rest of .\mechanize-0.2.5 you don't need.
| |
| #You can also see other places where you could put the "mechanize" folder. Write this in pymol to see the paths where pymol is searching for "mechanize"
| |
| # import sys; print(sys.path)
| |
| | |
| #-------------------------------------------------------------------------------
| |
| """
| |
| Example for pymol script to start the functions. For example: trypropka.pml
| |
| Execute with pymol or start pymol and: File->Run->trypropka.pml
| |
| ##############################################################################################################################################################################################################################
| |
| | |
| ### Point to your directory with your pdb file and where to save the results
| |
| #cd /homes/linnet/Documents/Speciale/5NT-project/Mutant-construct/predict_reactivity/propka
| |
| cd C:/Users/tlinnet/Documents/My Dropbox/Speciale/5NT-project/Mutant-construct/predict_reactivity/propka
| |
| | |
| ### If you have the script in your working directory the
| |
| #run ./propka.py
| |
| ### You can also make the script general available. Put it into your python path. Ex: C:\Program Files (x86)\PyMOL\PyMOL\modules Then do instead:
| |
| import propka
| |
| | |
| ### The fastest method is just to write propka. Then the last pymol molecule is assumed and send to server. verbose=yes makes the script gossip mode.
| |
| fetch 4ins, async=0
| |
| propka
| |
| #fetch 1hp1, async=0
| |
| #propka logtime=_, resi=5-10.20-30, resn=CYS.ATP.TRP, verbose=yes
| |
| | |
| ### Fetch 4ins from web. async make sure, we dont execute script before molecule is loaded. The resi and resn prints the interesting results right to command line.
| |
| #fetch 4ins, async=0
| |
| #propka chain=*, resi=5-10.20-30, resn=ASP.CYS, logtime=_
| |
| | |
| ### If there is no web connection, one can process a local .pka file. Either from a previous run or from a downloaded propka webpage result.
| |
| ### Then run and point to .pka file with: pkafile=./Results_propka/pkafile.pka Remember the dot "." in the start, to make it start in the current directory.
| |
| #load 4ins.pdb
| |
| #propka pkafile=./Results_propka/4ins_.pka, resi=18.25-30, resn=cys,
| |
| | |
| ### Some more examples. This molecule has 550 residues, so takes a longer time. We select to run the last molecule, by writing: molecule=1hp1
| |
| #fetch 4ins, async=0
| |
| #fetch 1hp1, async=0
| |
| #propka molecule=1hp1, chain=A, resi=300-308.513, resn=CYS.ATP.TRP, logtime=_, verbose=no, showresult=no
| |
| #propka molecule=1hp1, pkafile=./Results_propka/1hp1_.pka, verbose=yes
| |
| | |
| ### One can also just make a lookup for a protein. Use function: getpropka
| |
| ### Note. This does only print the result to the pymol command line
| |
| #getpropka source=ID, PDBID=4ake, logtime=_, showresult=yes
| |
| #getpropka source=ID, PDBID=4ins, logtime=_, server_wait=10.0, verbose=yes, showresult=no
| |
| ############################################Input parameters: propka############################################
| |
| ############# The order of input and changable things:
| |
| ############# propka(molecule="NIL",chain="*",resi="0",resn="NIL",method="upload",logtime=time.strftime("%m%d",time.localtime()),server_wait=3.0,version="v3.1",verbose="no",showresult="no",pkafile="NIL")
| |
| # method : method=upload is default. This sends .pdb file and request result from propka server.
| |
| ## method=file will only process a manual .pka file, and write a pymol command file. No use of mechanize.
| |
| ## If one points to an local .pka file, then method is auto-changed to method=file. This is handsome in off-line environment, ex. teaching or seminar.
| |
| # pkafile: Write the path to .pka file. Ex: pkafile=./Results_propka/4ins_.pka
| |
| # molecule : name of the molecule. Ending of file is assumed to be .pdb
| |
| # chain : which chains are saved to file, before molecule file is send to server. Separate with "." Ex: chain=A.b
| |
| # resi : Select by residue number, which residues should be printed to screen and saved to the log file: /Results_propka/_Results.log.
| |
| ## Separate with "." or make ranges with "-". Ex: resi=35.40-50
| |
| # resn : Select by residue name, which residues should be printed to screen and saved to the log file: /Results_propka/_Results.log.
| |
| ## Separate with "." Ex: resn=cys.tyr
| |
| # logtime : Each execution give a set of files with the job id=logtime. If logtime is not provided, the current time is used.
| |
| ## Normal it usefull to set it empty. Ex: logtime=_
| |
| # verbose : Verbose is switch, to turn on messages for the mechanize section. This is handsome to see how mechanize works, and for error searching.
| |
| # showresult : Switch, to turn on all results in pymol command window. Ex: showresult=yes
| |
| # server_wait=10.0 is default. This defines how long time between asking the server for a result. Set no lower than 3 seconds.
| |
| # version=v3.1 is default. This is what version of propka which would be used.
| |
| ## Possible: 'v3.1','v3.0','v2.0'. If a newer version is available than the current v3.1, a error message is raised to make user update the script.
| |
| ############################################Input parameters: getpropka############################################
| |
| ############# The order of input and changable things:
| |
| ############# getpropka(PDB="NIL",chain="*",resi="0",resn="NIL",source="upload",PDBID="",logtime=time.strftime("%Y%m%d%H%M%S",time.localtime()),server_wait=3.0,version="v3.1",verbose="no",showresult="no")
| |
| # PDB: points the path to a .pdb file. This is auto-set from propka function.
| |
| # source : source=upload is default and is set at the propka webpage.
| |
| # source=ID, PDBID=4ake , one can print to the command line, the pka value for any official pdb ID. No files are displayed in pymol.
| |
| # PDBID: is used as the 4 number/letter pdb code, when invoking source=ID.
| |
| | |
| ##############################################################################################################################################################################################################################
| |
| """
| |
| try: from pymol import cmd; runningpymol='yes'
| |
| except: runningpymol='no'; pass
| |
| import time, platform, os
| |
| | |
| def propka(molecule="NIL",chain="*",resi="0",resn="NIL",method="upload",logtime=time.strftime("%m%d",time.localtime()),server_wait=3.0,version="v3.1",verbose="no",showresult="no",pkafile="NIL",makebonds="yes"):
| |
| Script_Version="20110823"
| |
| ### First we have to be sure, we give reasonable arguments
| |
| if pkafile!="NIL":
| |
| method='file'
| |
| assert method in ['upload', 'file'], "'method' has to be either: method=upload or method=file"
| |
| ### If molecule="all", then try to get the last molecule
| |
| ##assert molecule not in ['NIL'], "You always have to provide molecule name. Example: molecule=4ins"
| |
| if molecule=="NIL":
| |
| assert len(cmd.get_names())!=0, "Did you forget to load a molecule? There are no objects in pymol."
| |
| molecule=cmd.get_names()[-1]
| |
| ### To print out to screen for selected residues. Can be separated with "." or make ranges with "-". Example: resi="4-8.10"
| |
| if resi != "0": resi_range = ResiRange(resi)
| |
| else: resi_range=[]
| |
| ### Also works for residue names. They are all converted to bigger letters. Example: resn="cys.Tyr"
| |
| if resn != "NIL": resn_range = ResnRange(resn)
| |
| else: resn_range = resn
| |
| ### Make chain range, and upper case.
| |
| chain = ChainRange(chain)
| |
| ### Make result directory. We also the absolut path to the new directory.
| |
| Newdir = createdirs()
| |
| if method=="upload":
| |
| ### We try to load mechanize. If this fail, one can always get the .pka file manual and the run: method=file
| |
| try: import mechanize; importedmechanize='yes'
| |
| except ImportError: print("Import error. Is a module missing?"); print(sys.exc_info()); print("Look if missing module is in your python path\n%s")%sys.path;importedmechanize='no'; import mechanize
| |
| ### The name for the new molecule
| |
| newmolecule = "%s%s"%(molecule,logtime)
| |
| ### Create the new molecule from original loaded and for the specified chains. Save it, and disable the old molecule.
| |
| cmd.create("%s"%newmolecule, "%s and chain %s"%(molecule,chain))
| |
| cmd.save("%s%s.pdb"%(Newdir,newmolecule), "%s"%newmolecule)
| |
| cmd.disable("%s"%molecule)
| |
| if molecule=="all": cmd.enable("%s"%molecule); cmd.show("cartoon", "%s"%molecule)
| |
| ### Let the new molecule be shown in cartoon.
| |
| cmd.hide("everything", "%s"%newmolecule)
| |
| cmd.show("cartoon", "%s"%newmolecule)
| |
| ### Make the absolut path to the newly created .pdb file.
| |
| PDB="%s%s.pdb"%(Newdir,newmolecule);source="upload"; PDBID=""
| |
| ### Request server, and get the absolut path to the result file.
| |
| pkafile = getpropka(PDB,chain,resi,resn,source,PDBID,logtime,server_wait,version,verbose,showresult)
| |
| ### Open the result file and put in into a handy list.
| |
| list_results,ligands_results = importpropkaresult(pkafile)
| |
| ### Now we check if the script is actually the newest one.
| |
| Web_Version,Script_Version=checkversion(Script_Version,verbose)
| |
| if float(Web_Version) > float(Script_Version):
| |
| print('\n\n####################################\nWarning: The author has updated the pymol propka script.\nPresent: %s > Script: %s \nThe new script is available at "http://pymolwiki.org/index.php/Propka" or "http://tinyurl.com/pymolpropka"\n####################################\n\n'%(Web_Version,Script_Version))
| |
| if method=="file":
| |
| assert pkafile not in ['NIL'], "You have to provide path to file. Example: pkafile=./Results_propka/4ins_2011.pka"
| |
| assert ".pka" in pkafile, 'The propka result file should end with ".pka" \nExample: pkafile=./Results_propka/4ins_2011.pka \npkafile=%s'%(pkafile)
| |
| ### The name for the molecule we pass to the writing script of pymol commands
| |
| newmolecule = "%s"%molecule
| |
| cmd.hide("everything", "%s"%newmolecule)
| |
| cmd.show("cartoon", "%s"%newmolecule)
| |
| ### We open the result file we have got in the manual way and put in into a handy list.
| |
| list_results,ligands_results = importpropkaresult(pkafile)
| |
| ### Then we print the interesting residues to the screen.
| |
| printpropkaresult(list_results, resi, resi_range, resn, resn_range, showresult, ligands_results)
| |
| ### Now create the pymol command file. This should label the protein. We get back the absolut path to the file, so we can execute it.
| |
| result_pka_pymol_name = writepymolcmd(newmolecule,pkafile,verbose,makebonds)
| |
| ### Now run our command file. But only if we are running pymol.
| |
| if runningpymol=='yes': cmd.do("run %s"%result_pka_pymol_name)
| |
| ##if runningpymol=='yes': cmd.do("@%s"%result_pka_pymol_name)
| |
| return(list_results)
| |
| if runningpymol !='no': cmd.extend("propka",propka)
| |
| | |
| def getpropka(PDB="NIL",chain="*",resi="0",resn="NIL",source="upload",PDBID="",logtime=time.strftime("%Y%m%d%H%M%S",time.localtime()),server_wait=3.0,version="v3.1",verbose="no",showresult="no"):
| |
| try: import mechanize; importedmechanize='yes'
| |
| except ImportError: print("Import error. Is a module missing?"); print(sys.exc_info()); print("Look if missing module is in your python path \n %s"%sys.path);importedmechanize='no'
| |
| propka_v_201108 = 3.1
| |
| url = "http://propka.ki.ku.dk/"
| |
| assert version in ['v2.0', 'v3.0', 'v3.1'], "'version' has to be either: 'v2.0', 'v3.0', 'v3.1'"
| |
| assert source in ['ID', 'upload', 'addr', 'input_file'], "'source' has to be either: 'ID', 'upload', 'addr', 'input_file'"
| |
| if source=="upload": assert PDB not in ['NIL'], "You always have to provide PDB path. Example: PDB=.\Results_propka\4ins2011.pdb"
| |
| if source=="ID": assert len(PDBID)==4 , "PDBID has to be 4 characters"
| |
| ### To print out to screen for selected residues. Can be separated with "." or make ranges with "-". Example: resi="4-8.10"
| |
| if resi != "0": resi_range = ResiRange(resi)
| |
| else: resi_range=[]
| |
| ### Also works for residue names. They are all converted to bigger letters. Example: resn="cys.Tyr"
| |
| if resn != "NIL": resn_range = ResnRange(resn)
| |
| else: resn_range = resn
| |
| ### Start the browser
| |
| br = mechanize.Browser()
| |
| ### We pass to the server, that we are not a browser, but this python script. Can be used for statistics at the propka server.
| |
| br.addheaders = [('User-agent', 'pythonMechanizeClient')]
| |
| ### To turn on debugging messages
| |
| ##br.set_debug_http(True)
| |
| ### To open the start page.
| |
| page_start = br.open(url)
| |
| read_start = page_start.read()
| |
| if verbose == 'yes': print(br.title()); print(br.geturl())
| |
| ### To get available forms
| |
| page_forms = [f.name for f in br.forms()]
| |
| if verbose == 'yes': print(page_forms)
| |
| ### Select first form
| |
| br.select_form(name=page_forms[0])
| |
| ## Print the current selected form, so we see that we values we start with.
| |
| if verbose == 'yes': print(br.form)
| |
| ### Print the parameters of the 'version' RadioControl button and current value
| |
| if verbose == 'yes': print(br.find_control(name='version')), br.find_control(name='version').value
| |
| ### This is to check, that the current script is "up-to-date".
| |
| propka_v_present = float(br.find_control(name='version').value[0].replace('v',''))
| |
| if propka_v_present > propka_v_201108:
| |
| raise UserWarning('\nNew version of propka exist.\nCheck/Update your script.\nPresent:v%s > Script:v%s'%(propka_v_present,propka_v_201108))
| |
| ### Change the parameters of the 'version' radio button and then reprint the new value. Input has to be in a list [input].
| |
| br.form['version'] = [version]
| |
| if verbose == 'yes': print(br.find_control(name='version').value)
| |
| ### Print the parameters of the 'source' RadioControl button and current value
| |
| if verbose == 'yes': print(br.find_control(name='source'), br.find_control(name='source').value)
| |
| ### Change the parameters of the 'source' radio button and then reprint the new value. Input has to be in a list [input].
| |
| br.form['source'] = [source]
| |
| if verbose == 'yes': print(br.find_control(name='source').value)
| |
| ### This step was the must strange and took a long time. For finding the information and the double negative way.
| |
| ### One have to enable the pdb button. Read more here: http://wwwsearch.sourceforge.net/old/ClientForm/ ("# All Controls may be disabled.....)
| |
| PDBID_control = br.find_control("PDBID")
| |
| PDB_control = br.find_control("PDB")
| |
| if verbose == 'yes': print(PDBID_control.disabled, PDB_control.disabled)
| |
| if source == "ID": PDBID_control.disabled=False; PDB_control.disabled=True
| |
| if source == "upload": PDBID_control.disabled=True; PDB_control.disabled=False
| |
| if verbose == 'yes': print(PDBID_control.disabled, PDB_control.disabled)
| |
| ### We create the result dir, and take with us the 'path' to the result dir.
| |
| Newdir = createdirs()
| |
| ### Open all the files, and assign them.
| |
| if source == "upload": filename = PDB
| |
| if source == "ID": filename = PDBID
| |
| files = openfiles(Newdir, filename, logtime, source)
| |
| result_pka_file=files[0];result_input_pka_file=files[1];result_log=files[2];filepath=files[3];result_pka_pkafile=files[4];result_pka_file_stripped=files[5];result_pka_file_bonds=files[6]
| |
| ## Print the parameters of the 'PDBID' TextControl button and current value
| |
| if source == "ID" and verbose == 'yes': print(br.find_control(name='PDBID')); print(br.find_control(name='PDBID').value)
| |
| ## Change the parameters of the 'PDBID' TextControl and then reprint the new value. Input has just to be a string.
| |
| if source == "ID": br.form["PDBID"] = PDBID
| |
| if source == "ID" and verbose == 'yes': print(br.find_control(name='PDBID').value)
| |
| ## Print the parameters of the 'PDB' TextControl button and current value
| |
| if source == "upload" and verbose == 'yes': print(br.find_control(name='PDB')); print(br.find_control(name='PDB').value)
| |
| ## Change the parameters of the 'PDB' FileControl and then reprint the new value. Input has just to be a string.
| |
| if source == "upload": PDBfilename=PDB; PDBfilenamepath=PDB
| |
| if source == "upload": br.form.add_file(open(PDBfilename), 'text/plain', PDBfilenamepath, name='PDB')
| |
| if source == "upload" and verbose == 'yes': print(br.find_control(name='PDB')); print(br.find_control(name='PDB').value)
| |
| ## Now reprint the current selected form, so we see that we have the right values.
| |
| if verbose == 'yes': print(br.form)
| |
| ### Make "how" we would like the next request. We would like to "Click the submit button", but we have not opened the request yet.
| |
| req = br.click(type="submit", nr=0)
| |
| ### Have to pass by a mechanize exception. Thats the reason for the why True
| |
| ### The error was due to: br.open(req)
| |
| ###### mechanize._response.httperror_seek_wrapper: HTTP Error refresh: The HTTP server returned a redirect error that would lead to an infinite loop.
| |
| ###### The last 30x error message was:
| |
| ###### OK
| |
| ### I haven't been able to find the refresh problem or extend the time. So we make a pass on the raised exception.
| |
| try:
| |
| print("Now sending request to server")
| |
| br.open(req)
| |
| ### If there is raised an exception, we jump through to the result page after some sleep.
| |
| except mechanize.HTTPError:
| |
| ### We can extract the jobid from the current browser url.
| |
| jobid = br.geturl()[32:-5]
| |
| ### We notice how the script at the server presents the final result page.
| |
| url_result = url + "pka/" + jobid + ".html"
| |
| ### Now we continue to try to find the result page, until we have succes. If page doesn't exist, we wait a little.
| |
| while True:
| |
| print("Result still not there. Waiting %s seconds more"%server_wait)
| |
| time.sleep(float(server_wait))
| |
| ### To pass the "break" after the exception, we make a hack, wait and then go to the result page, which is the jobid.
| |
| try:
| |
| page_result = br.open(url_result)
| |
| read_result = page_result.read()
| |
| ### If we don't receive a error in getting the result page, we break out of the while loop.
| |
| break
| |
| ### If the page doesn't exist yet. We go back in the while loop.
| |
| except mechanize.HTTPError:
| |
| ### Wait another round
| |
| pass
| |
| ### If we get a timeout, we also wait.
| |
| except mechanize.URLError:
| |
| ### Wait another round
| |
| pass
| |
| htmlresult="The detailed result is now available at: %s"%br.geturl()
| |
| print(htmlresult)
| |
| read_result = br.response().read()
| |
| ## Now save the available links from the current page. But only links that satisfy the expression.
| |
| links_result = []
| |
| for l in br.links(url_regex='http://propka.ki.ku.dk/pka'):
| |
| links_result.append(l)
| |
| ## We also extract the information for neighbour bons. This is given in the url links.
| |
| bonds=[]
| |
| for l in br.links(url_regex='http://propka.ki.ku.dk/view/new_view.cgi'):
| |
| l_split=str(l).split()
| |
| lresn=l_split[2]
| |
| lresi=l_split[3]
| |
| lchain=l_split[4]
| |
| lurl=l_split[1]
| |
| lurl_split=lurl.split("&")
| |
| lresn2=lurl_split[1]
| |
| lchain2=lurl_split[2]
| |
| lpka=lurl_split[3]
| |
| ldesolvation=lurl_split[4]
| |
| lneighbours=lurl_split[5:]
| |
| for i in range(len(lneighbours)):
| |
| bonds.append([lresn,lresi,lchain,lresn2,lchain2,lpka,ldesolvation,lneighbours[i]])
| |
| ### Now follow the link to the .propka_input resultpage
| |
| if len(links_result) > 1: br.follow_link(links_result[1])
| |
| ### Now get the page text for the current link
| |
| if len(links_result) > 1: read_result1 = br.response().read()
| |
| ### Save the result
| |
| if len(links_result) > 1: result_input_pka_file.write(read_result1)
| |
| ### Now follow the link to the .pka resultpage
| |
| if len(links_result) > 1: br.back()
| |
| result_input_pka_file.close()
| |
| ### Now follow first link. "Should be" available for all versions of propka.
| |
| br.follow_link(links_result[0])
| |
| ### Now get the page for the current link
| |
| read_result0 = br.response().read()
| |
| ### Save the result and close file.
| |
| result_pka_file.write(read_result0)
| |
| result_pka_file.close()
| |
| ### Now get the result in a list, which is sorted
| |
| list_results,ligands_results = importpropkaresult(result_pka_pkafile)
| |
| ### Print to log file
| |
| result_log.write("# executed: %s \n# logtime: %s \n# source=%s \n# PDB=%s \n# chain=%s \n# PDBID=%s \n# server_wait=%s version=%s verbose=%s showresult=%s \n# resi=%s resn=%s\n# %s \n"%(time.strftime("%Y%m%d%H%M%S",time.localtime()),logtime,source,PDB,chain,PDBID,server_wait,version,verbose,showresult,resi,resn,htmlresult))
| |
| ### Print to screen
| |
| printpropkaresult(list_results, resi, resi_range, resn, resn_range, showresult, ligands_results)
| |
| ### Now write to log and the stripped file
| |
| for l in list_results:
| |
| if resi != "0" and int(l[1]) in resi_range:
| |
| result_log.write("%3s %3s %s %6s %3s %5s %3s %4s %s"%(l[0],l[1],l[2],l[3],l[4],l[5],l[6],l[7],l[8]) + '\n')
| |
| if resn != "NIL" and l[0] in resn_range and int(l[1]) not in resi_range:
| |
| result_log.write("%3s %3s %s %6s %3s %5s %3s %4s %s"%(l[0],l[1],l[2],l[3],l[4],l[5],l[6],l[7],l[8]) + '\n')
| |
| result_pka_file_stripped.write("%3s %3s %s %6s %3s %5s %3s %4s %s"%(l[0],l[1],l[2],l[3],l[4],l[5],l[6],l[7],l[8]) + '\n')
| |
| for l in ligands_results:
| |
| if resn != "NIL" and l[0] in resn_range:
| |
| result_log.write("%3s %3s %s %6s %3s %5s %3s %4s %s"%(l[0],l[1],l[2],l[3],l[4],l[5],l[6],l[7],l[8]) + '\n')
| |
| result_pka_file_stripped.write("%3s %3s %s %6s %3s %5s %3s %4s %s"%(l[0],l[1],l[2],l[3],l[4],l[5],l[6],l[7],l[8]) + '\n')
| |
| result_pka_file_stripped.close()
| |
| result_log.close()
| |
| ### Now handle the bonds. We have to delete dublicates first.
| |
| bonds.sort()
| |
| last=bonds[-1]
| |
| for i in range(len(bonds)-2, -1, -1):
| |
| if last == bonds[i]: del bonds[i]
| |
| else: last=bonds[i]
| |
| ### Now make a selection for known residue
| |
| bonds_selected=[]
| |
| bonds_ligands=[]
| |
| for l in bonds:
| |
| if l[0][6:] in ['ASP', 'GLU', 'ARG', 'LYS', 'HIS', 'CYS', 'TYR', 'C-', 'N+']:
| |
| bonds_selected.append(l)
| |
| else:
| |
| bonds_ligands.append(l)
| |
| ### And now sort it.
| |
| bonds_selected.sort(key=lambda residue: int(residue[1]))
| |
| ### Now write it to file
| |
| bonddic={'=':' ',':':' ',',':' ',"'":" "}
| |
| for l in bonds_selected:
| |
| nb = replace_all(l[7],bonddic)
| |
| result_pka_file_bonds.write("%3s %3s %s %7s %7s %9s %17s %s"%(l[0][6:],l[1],l[2][:1],l[3][8:],l[4],l[5],l[6],nb) + '\n')
| |
| for l in bonds_ligands:
| |
| nb = replace_all(l[7],bonddic)
| |
| result_pka_file_bonds.write("%3s %3s %s %7s %7s %9s %17s %s"%(l[0][6:],l[1],l[2][:1],l[3][8:],l[4],l[5],l[6],nb) + '\n')
| |
| result_pka_file_bonds.close()
| |
| return(result_pka_pkafile)
| |
| if runningpymol !='no': cmd.extend("getpropka",getpropka)
| |
| | |
| def openpymolfiles(pkafile):
| |
| result_pka_pymol_name = pkafile.replace(".pka",".pml")
| |
| result_pka_pymol = open(result_pka_pymol_name, "w")
| |
| return(result_pka_pymol, result_pka_pymol_name)
| |
| | |
| def printpropkaresult(list_results, resi, resi_range, resn, resn_range, showresult, ligands_results):
| |
| for l in list_results:
| |
| if resi != "0" and int(l[1]) in resi_range:
| |
| if showresult != 'yes': print("%3s %3s %s %6s %3s %5s %3s %4s %s"%(l[0],l[1],l[2],l[3],l[4],l[5],l[6],l[7],l[8]))
| |
| if resn != "NIL" and l[0] in resn_range and int(l[1]) not in resi_range:
| |
| if showresult != 'yes': print("%3s %3s %s %6s %3s %5s %3s %4s %s"%(l[0],l[1],l[2],l[3],l[4],l[5],l[6],l[7],l[8]))
| |
| if showresult == 'yes': print("%3s %3s %s %6s %3s %5s %3s %4s %s"%(l[0],l[1],l[2],l[3],l[4],l[5],l[6],l[7],l[8]))
| |
| for l in ligands_results:
| |
| if resn != "NIL" and l[0] in resn_range:
| |
| if showresult != 'yes': print("%3s %3s %s %6s %3s %5s %3s %4s %s"%(l[0],l[1],l[2],l[3],l[4],l[5],l[6],l[7],l[8]))
| |
| if showresult == 'yes': print("%3s %3s %s %6s %3s %5s %3s %4s %s"%(l[0],l[1],l[2],l[3],l[4],l[5],l[6],l[7],l[8]))
| |
| | |
| def importpropkaresult(result_pka_pkafile):
| |
| result_pka_file = open(result_pka_pkafile, "r")
| |
| list_results = []
| |
| ligands_results = []
| |
| ##bonding_partners = []
| |
| for l in result_pka_file:
| |
| if not l.strip():
| |
| continue
| |
| else:
| |
| ### To search for the right lines
| |
| if l.strip().split()[0] in ['ASP', 'GLU', 'ARG', 'LYS', 'HIS', 'CYS', 'TYR', 'C-', 'N+'] and len(l.strip().split())>20:
| |
| list_results.append([l.strip().split()[0], l.strip().split()[1], l.strip().split()[2], l.strip().split()[3], l.strip().split()[4], l.strip().split()[6], l.strip().split()[7], l.strip().split()[8], l.strip().split()[9]])
| |
| ##bonding_partners.append(l.strip().split()[11]);bonding_partners.append(l.strip().split()[15]);bonding_partners.append(l.strip().split()[19])
| |
| if l.strip().split()[0] not in ['ASP', 'GLU', 'ARG', 'LYS', 'HIS', 'CYS', 'TYR', 'C-', 'N+'] and len(l.strip().split())>20:
| |
| ligands_results.append([l.strip().split()[0], l.strip().split()[1], l.strip().split()[2], l.strip().split()[3], l.strip().split()[4], l.strip().split()[6], l.strip().split()[7], l.strip().split()[8], l.strip().split()[9]])
| |
| ##bonding_partners.append(l.strip().split()[11]);bonding_partners.append(l.strip().split()[15]);bonding_partners.append(l.strip().split()[19])
| |
| ### Sort the result after the residue number and then chain.
| |
| list_results.sort(key=lambda residue: int(residue[1]))
| |
| list_results.sort(key=lambda chain: chain[2])
| |
| ##bonding_partners=uniqifi(bonding_partners)
| |
| ##bonding_partners[:] = [x for x in bonding_partners if x != "XXX"]
| |
| result_pka_file.close()
| |
| return(list_results,ligands_results)
| |
| | |
| def importpropkabonds(result_pka_pkafile):
| |
| bonds=[]
| |
| result_pka_file_bonds=open(result_pka_pkafile[:-4]+".bonds", "r")
| |
| for l in result_pka_file_bonds:
| |
| bonds.append(l.split())
| |
| result_pka_file_bonds.close()
| |
| return(bonds)
| |
| | |
| def createdirs():
| |
| if platform.system() == 'Windows': Newdir = os.getcwd()+"\Results_propka\\"
| |
| if platform.system() == 'Linux': Newdir = os.getcwd()+"/Results_propka/"
| |
| if not os.path.exists(Newdir): os.makedirs(Newdir)
| |
| return(Newdir)
| |
| | |
| def openfiles(Newdir, filename, logtime, source):
| |
| if source == "upload":
| |
| result_pka_pkafile = filename.replace(".pdb",".pka")
| |
| result_pka_input_pkafile = filename.replace(".pdb",".propka_input")
| |
| result_log_name = "%s_Results.log"%(Newdir)
| |
| result_pka_file_stripped_name = filename.replace(".pdb",".stripped")
| |
| result_pka_file_bonds_name = filename.replace(".pdb",".bonds")
| |
| if source == "ID":
| |
| result_pka_pkafile = "%s%s%s.pka"%(Newdir,filename,logtime)
| |
| result_pka_input_pkafile = "%s%s%s.propka_input"%(Newdir,filename,logtime)
| |
| result_log_name = "%s_Results.log"%(Newdir)
| |
| result_pka_file_stripped_name = "%s%s%s.stripped"%(Newdir,filename,logtime)
| |
| result_pka_file_bonds_name = "%s%s%s.bonds"%(Newdir,filename,logtime)
| |
| if platform.system() == 'Windows': filepath = "\\"
| |
| if platform.system() == 'Linux': filepath = "/"
| |
| ### Open the files
| |
| result_pka_file = open(result_pka_pkafile, "w")
| |
| result_input_pka_file = open(result_pka_input_pkafile, "w")
| |
| result_log = open(result_log_name, "a")
| |
| result_pka_file_stripped = open(result_pka_file_stripped_name, "w")
| |
| result_pka_file_bonds = open(result_pka_file_bonds_name, "w")
| |
| return(result_pka_file, result_input_pka_file, result_log, filepath, result_pka_pkafile,result_pka_file_stripped,result_pka_file_bonds)
| |
| | |
| def ResiRange(resi):
| |
| resi = resi.split('.')
| |
| resiList = []
| |
| for i in resi:
| |
| if '-' in i:
| |
| tmp = i.split('-')
| |
| resiList.extend(range(int(tmp[0]),int(tmp[-1])+1))
| |
| if '-' not in i:
| |
| resiList.append(int(i))
| |
| return(resiList)
| |
| | |
| def ResnRange(resn):
| |
| resn_split = resn.split('.')
| |
| resn_range = [resnr.upper() for resnr in resn_split]
| |
| return(resn_range)
| |
| | |
| def ChainRange(chain):
| |
| chainstring = chain.replace(".","+").upper()
| |
| return(chainstring)
| |
| | |
| def writepymolcmd(newmolecule,pkafile,verbose,makebonds):
| |
| list_results,ligands_results = importpropkaresult(pkafile)
| |
| ### Now find the available bonding partners that pymol knows of
| |
| bonding_partners = []
| |
| bonding_partners_str = cmd.get_pdbstr("%s and resn * and not resn ASP+GLU+ARG+LYS+HIS+CYS+TYR+GLN+ASN+SER+THR+GLY+PHE+LEU+ALA+ILE+TRP+MET+PRO+VAL+HOH"%(newmolecule))
| |
| for i in range(len(bonding_partners_str.splitlines())-1):
| |
| bonding_partners_split = bonding_partners_str.splitlines()[i].split()
| |
| if bonding_partners_split[0] == "HETATM" or bonding_partners_split[0] == "ATOM" :
| |
| bonding_partners_single = bonding_partners_split[3]
| |
| bonding_partners.append(bonding_partners_single)
| |
| bonding_partners=uniqifi(bonding_partners)
| |
| if verbose == 'yes': print("And other possible bonding partners is: %s"%(bonding_partners))
| |
| ### Read in the bond file, if it exists
| |
| writebonds="no"
| |
| if os.path.isfile(pkafile[:-4]+".bonds") and makebonds == "yes": bonds = importpropkabonds(pkafile); writebonds="yes"
| |
| ### Open the pymol command file for writing
| |
| files_pka_pymol = openpymolfiles(pkafile)
| |
| result_pka_pymol=files_pka_pymol[0];result_pka_pymol_name=files_pka_pymol[1]
| |
| ### Make some dictionary for propka->pymol name conversion
| |
| dictio = {'ASP':'CG', 'GLU':'CD', 'ARG':'CZ', 'LYS':'NZ', 'HIS':'CG', 'CYS':'SG', 'TYR':'OH', 'C-':'C', 'N+':'N','NTR':'N','CTR':'C','GLN':'CD','ASN':'CG','SER':'OG','THR':'OG1','GLY':'CA','PHE':'CZ','LEU':'CG','ALA':'CB','ILE':'CD1','TRP':'NE1','MET':'SD','PRO':'CG','VAL':'CB'}
| |
| dictio2 = {'ASP':'D', 'GLU':'E', 'ARG':'R', 'LYS':'K', 'HIS':'H', 'CYS':'C', 'TYR':'Y', 'C-':'C-', 'N+':'N+'}
| |
| ### This list is from: http://en.wikipedia.org/wiki/Protein_pKa_calculations
| |
| pkaaminoacid=['ASP','GLU','ARG','LYS','HIS','CYS','TYR']
| |
| pkadictio = {'ASP':3.9, 'GLU':4.3, 'ARG':12.0, 'LYS':10.5, 'HIS':6.0, 'CYS':8.3, 'TYR':10.1}
| |
| ### Now start write to the file.
| |
| ### Try to make silent
| |
| ##result_pka_pymol.write("cmd.feedback('disable','all','actions')\n")
| |
| ##result_pka_pymol.write("cmd.feedback('disable','all','results')\n")
| |
| ### Change the GUI width, to make the long names possible.
| |
| result_pka_pymol.write("cmd.set('internal_gui_width','360')\n")
| |
| ### Set fonts
| |
| result_pka_pymol.write("cmd.set('label_font_id','12')\n")
| |
| result_pka_pymol.write("cmd.set('label_size','-0.5')\n")
| |
| result_pka_pymol.write("cmd.set('label_color','grey')\n")
| |
| ### No auto zoom the new objects
| |
| result_pka_pymol.write("cmd.set('auto_zoom','off')\n")
| |
| ### The name for the molecules are defined here
| |
| pkamolecule="%spKa"%(newmolecule)
| |
| pkalabelmolecule="%sLab"%(newmolecule)
| |
| ### Create the groups now, so they come in order. They will be empty
| |
| result_pka_pymol.write("cmd.group('%sResi','Res*')\n"%(newmolecule))
| |
| result_pka_pymol.write("cmd.group('%sLigands','Lig*')\n"%(newmolecule))
| |
| if writebonds=="yes": result_pka_pymol.write("cmd.group('%sBonds','%sBond*')\n"%(newmolecule,newmolecule))
| |
| ### Create new empty pymol pka molecules. For pka atoms and its label. This is a "bucket" we where we will put in the atoms together.
| |
| result_pka_pymol.write("cmd.create('%s','None')\n"%(pkamolecule))
| |
| result_pka_pymol.write("cmd.create('%s','None')\n"%(pkalabelmolecule))
| |
| ### Now make the pka atoms and alter, color and such
| |
| for l in list_results:
| |
| name=dictio[l[0]];resn=dictio2[l[0]];resi=l[1];chain=l[2];pka=l[3];buried=l[4]
| |
| if "*" in pka: pka = pka.replace("*",""); comment="*Coupled residue"
| |
| else: comment=""
| |
| if l[0] in pkaaminoacid:
| |
| pkadiff =(float(pka)-pkadictio[l[0]])
| |
| pkadiff = "(%s)"%pkadiff
| |
| if pka=="99.99": pkadiff=""
| |
| else: pkadiff=""
| |
| ### Make the selection for which atom to copy
| |
| newselection = ("/%s//%s/%s/%s"%(newmolecule,chain,resi,name))
| |
| protselect = ("%sRes_%s%s%s"%(newmolecule,chain,resn,resi))
| |
| result_pka_pymol.write("cmd.select('%s','byres %s')\n"%(protselect,newselection))
| |
| result_pka_pymol.write("cmd.show('sticks','byres %s')\n"%(protselect))
| |
| ### The temporary name
| |
| tempname = ("%s%s%s%s"%(pkamolecule,chain,resi,name))
| |
| tempnamelabel = ("%s%s%s%s"%(pkalabelmolecule,chain,resi,name))
| |
| tempselect= ("/%s//%s/%s"%(tempname,chain,resi))
| |
| tempselectlabel= ("/%s//%s/%s"%(tempnamelabel,chain,resi))
| |
| ### Copy the atom, call it by the residue name
| |
| result_pka_pymol.write("cmd.create('%s','%s',quiet=1)\n"%(tempname,newselection))
| |
| ### Alter the name and the b value of the newly created atom
| |
| result_pka_pymol.write("cmd.alter('%s','b=%s')\n"%(tempselect,pka))
| |
| result_pka_pymol.write("cmd.alter('%s','vdw=0.5')\n"%(tempselect))
| |
| result_pka_pymol.write("cmd.alter('%s','name=%s%s%s')\n"%(tempselect,'"',pka,'"'))
| |
| ### Now create a fake label atom, and translate it
| |
| result_pka_pymol.write("cmd.create('%s','%s',quiet=1)\n"%(tempnamelabel,tempname))
| |
| movelabelxyz = (1.5,0,0)
| |
| result_pka_pymol.write("cmd.translate('[%s,%s,%s]','%s',camera=0)\n"%(movelabelxyz[0],movelabelxyz[1],movelabelxyz[2],tempnamelabel))
| |
| ### Labelling alternate positions are not allowed, so we delete that attribute for the label atoms.
| |
| result_pka_pymol.write("cmd.alter('%s','alt=%s%s')\n"%(tempselectlabel,'"','"'))
| |
| result_pka_pymol.write("cmd.label('%s','text_type=%spka=%s%s Bu:%s%s%s%s')\n"%(tempselectlabel,'"',pka,pkadiff,buried,'%',comment,'"'))
| |
| ### Now put the atoms into a bucket of atoms
| |
| result_pka_pymol.write("cmd.create('%s','%s or (%s)',quiet=1)\n"%(pkamolecule,pkamolecule,tempselect))
| |
| result_pka_pymol.write("cmd.create('%s','%s or (%s)',quiet=1)\n"%(pkalabelmolecule,pkalabelmolecule,tempselectlabel))
| |
| ### Remove the temporary atoms
| |
| result_pka_pymol.write("cmd.remove('%s')\n"%(tempname))
| |
| result_pka_pymol.write("cmd.remove('%s')\n"%(tempnamelabel))
| |
| ### Delete the temporary molecule/selection
| |
| result_pka_pymol.write("cmd.delete('%s')\n"%(tempname))
| |
| result_pka_pymol.write("cmd.delete('%s')\n"%(tempnamelabel))
| |
| ### Group the resi together
| |
| result_pka_pymol.write("cmd.group('%sResi','%sRes*')\n"%(newmolecule,newmolecule))
| |
| for l in ligands_results:
| |
| resn=l[0];atom=l[1];chain=l[2];pka=l[3];buried=l[4]
| |
| if verbose == 'yes': print("Ligand. resn:%s atom:%s chain:%s pka:%s buried:%s"%(resn,atom,chain,pka,buried))
| |
| if Check_bonding_partners(bonding_partners, resn)[0]:
| |
| if "*" in pka: pka = pka.replace("*",""); comment="*Coupled residue"
| |
| else: comment=""
| |
| ### Make the selection for which atom to copy
| |
| ligselection = ("/%s and chain %s and resn %s and name %s"%(newmolecule,chain,resn,atom))
| |
| ligselect = ("%sLig_%s%s%s"%(newmolecule,chain,resn,atom))
| |
| result_pka_pymol.write("cmd.select('%s','%s')\n"%(ligselect,ligselection))
| |
| result_pka_pymol.write("cmd.show('sticks','byres %s')\n"%(ligselect))
| |
| result_pka_pymol.write("cmd.util.cbap('byres %s')\n"%(ligselect))
| |
| ### The temporary name
| |
| tempname = ("%s%s%s%s"%(pkamolecule,chain,resn,atom))
| |
| tempnamelabel = ("%s%s%s%s"%(pkalabelmolecule,chain,resn,atom))
| |
| tempselect= ("/%s and chain %s and resn %s"%(tempname,chain,resn))
| |
| tempselectlabel= ("/%s and chain %s and resn %s"%(tempnamelabel,chain,resn))
| |
| ### Copy the atom, call it by the residue name
| |
| result_pka_pymol.write("cmd.create('%s','%s',quiet=1)\n"%(tempname,ligselection))
| |
| ### Alter the name and the b value of the newly created atom
| |
| result_pka_pymol.write("cmd.alter('%s','b=%s')\n"%(tempselect,pka))
| |
| result_pka_pymol.write("cmd.alter('%s','vdw=0.5')\n"%(tempselect))
| |
| result_pka_pymol.write("cmd.alter('%s','name=%s%s%s')\n"%(tempselect,'"',pka,'"'))
| |
| ### Now create a fake label atom, and translate it
| |
| result_pka_pymol.write("cmd.create('%s','%s',quiet=1)\n"%(tempnamelabel,tempname))
| |
| movelabelxyz = (1.5,0,0)
| |
| result_pka_pymol.write("cmd.translate('[%s,%s,%s]','%s',camera=0)\n"%(movelabelxyz[0],movelabelxyz[1],movelabelxyz[2],tempnamelabel))
| |
| ### Labelling alternate positions are not allowed, so we delete that attribute for the label atoms.
| |
| result_pka_pymol.write("cmd.alter('%s','alt=%s%s')\n"%(tempselectlabel,'"','"'))
| |
| result_pka_pymol.write("cmd.label('%s','text_type=%spka=%s Bu:%s%s%s%s')\n"%(tempselectlabel,'"',pka,buried,'%',comment,'"'))
| |
| ### Now put the atoms into a bucket of atoms
| |
| result_pka_pymol.write("cmd.create('%s','%s or (%s)',quiet=1)\n"%(pkamolecule,pkamolecule,tempselect))
| |
| result_pka_pymol.write("cmd.create('%s','%s or (%s)',quiet=1)\n"%(pkalabelmolecule,pkalabelmolecule,tempselectlabel))
| |
| ### Remove the temporary atoms
| |
| result_pka_pymol.write("cmd.remove('%s')\n"%(tempname))
| |
| result_pka_pymol.write("cmd.remove('%s')\n"%(tempnamelabel))
| |
| ### Delete the temporary molecule/selection
| |
| result_pka_pymol.write("cmd.delete('%s')\n"%(tempname))
| |
| result_pka_pymol.write("cmd.delete('%s')\n"%(tempnamelabel))
| |
| ### Group the resi together
| |
| result_pka_pymol.write("cmd.group('%sLigands','%sLig*')\n"%(newmolecule,newmolecule))
| |
| ### Finish the pka atoms, and show spheres
| |
| result_pka_pymol.write("cmd.show('spheres','%s')\n"%(pkamolecule))
| |
| result_pka_pymol.write("cmd.spectrum('b','red_white_blue',selection='%s',minimum='0',maximum='14')\n"%(pkamolecule))
| |
| result_pka_pymol.write("cmd.alter('%s and name 99.9','vdw=0.8')\n"%(pkamolecule))
| |
| result_pka_pymol.write("cmd.show('spheres','%s and name 99.9')\n"%(pkamolecule))
| |
| result_pka_pymol.write("cmd.color('sulfur','%s and name 99.9')\n"%(pkamolecule))
| |
| ### Now we make the bonds
| |
| if writebonds=="yes":
| |
| Bondgroups=[]
| |
| naturalaminoacids = ['ASP','GLU','ARG','LYS','HIS','CYS','TYR','NTR','N+','CTR','C-','GLN','ASN','SER','THR','GLY','PHE','LEU','ALA','ILE','TRP','MET','PRO','VAL']
| |
| for l in bonds:
| |
| if l[0] in naturalaminoacids:
| |
| name=dictio[l[0]];resi=l[1];chain=l[2];desolvation=l[6][12:];pkachange=l[11];NBresi=l[8][3:];NBchain=l[9];NBbond=l[-1][:2]
| |
| if l[8][:3] in naturalaminoacids:
| |
| NBname,cutoff=BondTypeName(dictio[l[8][:3]],NBbond)
| |
| fromselection = ("/%s//%s/%s/%s"%(newmolecule,chain,resi,name))
| |
| toselection = ("/%s//%s/%s/%s"%(newmolecule,NBchain,NBresi,NBname))
| |
| if l[8][:3]=='NTR':
| |
| extind=cmd.identify("chain %s and name N"%(NBchain))[0]
| |
| toselection = ("/%s and id %s and name N"%(newmolecule,extind))
| |
| NBresi="N+"
| |
| if l[8][:3]=='CTR':
| |
| extind=cmd.identify("chain %s and name C"%(NBchain))[-1]
| |
| toselection = ("/%s and id %s and name C"%(newmolecule,extind))
| |
| NBresi="C-"
| |
| distname = ("%s_%s%s%s%s_%s_%s"%(newmolecule,chain,resi,NBchain,NBresi,NBbond,pkachange))
| |
| result_pka_pymol.write("cmd.distance('%s','%s','%s'%s)\n"%(distname,fromselection,toselection,cutoff))
| |
| result_pka_pymol.write("cmd.color('%s','%s')\n"%(SetDashColor(NBbond),distname))
| |
| ##result_pka_pymol.write("cmd.disable('%s')\n"%(distname))
| |
| Bondgroups.append("%s%s"%(chain,resi))
| |
| if l[8][:3] not in naturalaminoacids and Check_bonding_partners(bonding_partners, l[8])[0]:
| |
| cutoff=""; NBresn = Check_bonding_partners(bonding_partners, l[8])[1]; NBname=l[8][len(NBresn):]+"*"
| |
| fromselection = ("/%s//%s/%s/%s"%(newmolecule,chain,resi,name))
| |
| toselection = ("/%s and chain %s and resn %s and name %s"%(newmolecule,NBchain,NBresn,NBname))
| |
| if verbose == 'yes': print("Res->Ligand: (%s) -> (%s)"%(fromselection, toselection))
| |
| result_pka_pymol.write("cmd.show('sticks','%s')\n"%(toselection))
| |
| distname = ("%s_%s%s%s_%s_%s"%(newmolecule,chain,resi,NBresn,NBbond,pkachange))
| |
| result_pka_pymol.write("cmd.distance('%s','%s','%s'%s)\n"%(distname,fromselection,toselection,cutoff))
| |
| result_pka_pymol.write("cmd.color('%s','%s')\n"%(SetDashColor(NBbond),distname))
| |
| ##result_pka_pymol.write("cmd.disable('%s')\n"%(distname))
| |
| Bondgroups.append("%s%s"%(chain,resi))
| |
| if l[0] in bonding_partners:
| |
| resn=l[0];atom=l[1];chain=l[2];desolvation=l[6][12:];pkachange=l[11];NBresi=l[8][3:];NBchain=l[9];NBbond=l[-1][:2]
| |
| if not Check_bonding_partners(bonding_partners, l[8])[0]:
| |
| NBname,cutoff=BondTypeName(dictio[l[8][:3]],NBbond)
| |
| fromselection = ("/%s and chain %s and resn %s and name %s"%(newmolecule,chain,resn,atom))
| |
| toselection = ("/%s//%s/%s/%s"%(newmolecule,NBchain,NBresi,NBname))
| |
| if l[8][:3]=='NTR':
| |
| extind=cmd.identify("chain %s and name N"%(NBchain))[0]
| |
| toselection = ("/%s and id %s and name N"%(newmolecule,extind))
| |
| NBresi="N+"
| |
| if l[8][:3]=='CTR':
| |
| extind=cmd.identify("chain %s and name C"%(NBchain))[-1]
| |
| toselection = ("/%s and id %s and name C"%(newmolecule,extind))
| |
| NBresi="C-"
| |
| distname = ("%s_%s%s%s%s%s_%s_%s"%(newmolecule,chain,resn,atom,NBchain,NBresi,NBbond,pkachange))
| |
| result_pka_pymol.write("cmd.distance('%s','%s','%s'%s)\n"%(distname,fromselection,toselection,cutoff))
| |
| result_pka_pymol.write("cmd.color('%s','%s')\n"%(SetDashColor(NBbond),distname))
| |
| Bondgroups.append("%s%s%s"%(chain,resn,atom))
| |
| ##result_pka_pymol.write("cmd.disable('%s')\n"%(distname))
| |
| if Check_bonding_partners(bonding_partners, l[8])[0]:
| |
| cutoff=""; NBresn = Check_bonding_partners(bonding_partners, l[8])[1]; NBname=l[8][len(NBresn):]+"*"
| |
| fromselection = ("/%s and chain %s and resn %s and name %s"%(newmolecule,chain,resn,atom))
| |
| toselection = ("/%s and chain %s and resn %s and name %s"%(newmolecule,NBchain,NBresn,NBname))
| |
| if verbose == 'yes': print("Ligand->Ligand: (%s) -> (%s)"%(fromselection, toselection))
| |
| result_pka_pymol.write("cmd.show('sticks','%s')\n"%(toselection))
| |
| distname = ("%s_%s%s%s%s_%s_%s"%(newmolecule,chain,resn,atom,NBresn,NBbond,pkachange))
| |
| result_pka_pymol.write("cmd.distance('%s','%s','%s'%s)\n"%(distname,fromselection,toselection,cutoff))
| |
| result_pka_pymol.write("cmd.color('%s','%s')\n"%(SetDashColor(NBbond),distname))
| |
| ##result_pka_pymol.write("cmd.disable('%s')\n"%(distname))
| |
| Bondgroups.append("%s%s%s"%(chain,resn,atom))
| |
| Bondgroups=uniqifi(Bondgroups)
| |
| for l in Bondgroups:
| |
| result_pka_pymol.write("cmd.group('%sBonds_%s','%s_%s*')\n"%(newmolecule,l,newmolecule,l))
| |
| result_pka_pymol.write("cmd.disable('%sBonds_%s')\n"%(newmolecule,l))
| |
| result_pka_pymol.write("cmd.group('%sBonds','%sBonds_*')\n"%(newmolecule,newmolecule))
| |
| result_pka_pymol.write("cmd.set('auto_zoom','on')\n")
| |
| ##result_pka_pymol.write("cmd.feedback('enable','all','actions')\n")
| |
| ##result_pka_pymol.write("cmd.feedback('enable','all','results')\n")
| |
| result_pka_pymol.close()
| |
| return(result_pka_pymol_name)
| |
| | |
| def checkversion(Script_Version="0",verbose='no',url="http://pymolwiki.org/index.php/Propka#ScriptVersion"):
| |
| try: import mechanize; importedmechanize='yes'
| |
| except ImportError: print("Import error. Is a module missing?"); print(sys.exc_info()); print("Look if missing module is in your python path \n %s"%sys.path);importedmechanize='no'
| |
| ### Start the browser
| |
| br = mechanize.Browser()
| |
| ### We pass to the server, that we are not a browser, but this python script. Can be used for statistics for the server.
| |
| br.addheaders = [('User-agent', 'pythonMechanizeClient')]
| |
| ### To open the start page.
| |
| page_start = br.open(url)
| |
| read_start = page_start.read()
| |
| if verbose == 'yes': print(br.title()); print(br.geturl())
| |
| read_start_lines = read_start.splitlines()
| |
| for l in read_start_lines:
| |
| if "Current_Version=" in l:
| |
| Web_Version=l.split("=")[-1]
| |
| if verbose == 'yes': print(Web_Version)
| |
| return(Web_Version, Script_Version)
| |
| | |
| def replace_all(text, dic):
| |
| for i, j in dic.iteritems():
| |
| text = text.replace(i, j)
| |
| return(text)
| |
| | |
| def uniqifi(seq, idfun=None):
| |
| ### Order preserving
| |
| if idfun is None:
| |
| def idfun(x): return x
| |
| seen = {}
| |
| result = []
| |
| for item in seq:
| |
| marker = idfun(item)
| |
| if marker in seen: continue
| |
| seen[marker] = 1
| |
| result.append(item)
| |
| return(result)
| |
| | |
| def BondTypeName(NBname, NBbond):
| |
| if NBbond=="SH":
| |
| cutoff=""
| |
| return(NBname,cutoff)
| |
| if NBbond=="BH":
| |
| cutoff=""
| |
| return("N",cutoff)
| |
| else:
| |
| cutoff=""
| |
| return(NBname,cutoff)
| |
| | |
| def Check_bonding_partners(bonding_partners, NBname):
| |
| answer = False
| |
| for l in bonding_partners:
| |
| if l in NBname:
| |
| answer = True
| |
| NBname = l
| |
| break
| |
| else:
| |
| answer = False
| |
| return(answer,NBname)
| |
| | |
| def SetDashColor(NBbond):
| |
| if NBbond=="SH": color="brightorange"
| |
| if NBbond=="BH": color="lightorange"
| |
| if NBbond=="CC": color="red"
| |
| return(color)
| |
| </syntaxhighlight> | | </syntaxhighlight> |
|
| |
|