Surface: Difference between revisions
mNo edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
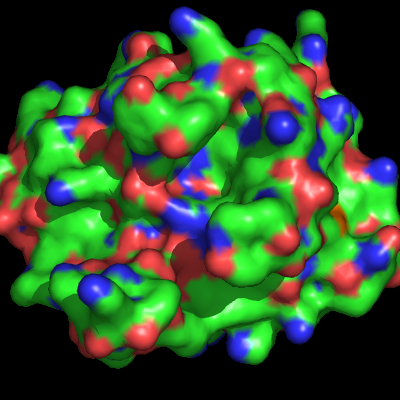
[[Image:Surface_ex.png|thumb|Surface Representation Example|right]] | The surface representation of a protein, in PyMol, shows the "Connolly" surface or the surface that would be traced out by the '''surfaces''' of waters in contact with teh protein at all possible positions. [[Image:Surface_ex.png|thumb|Surface Representation Example|right]] | ||
==Enabling== | |||
To enable the surface representation do | |||
show surface, SEL | |||
for any proper selection SEL. | |||
==Settings== | |||
===Transparency=== | |||
== | |||
To adjust the transparency of surfaces try: | To adjust the transparency of surfaces try: | ||
<source lang="python"> | <source lang="python"> | ||
| Line 13: | Line 15: | ||
Where 1.0 will be an invisible and 0.0 a completely solid surface. | Where 1.0 will be an invisible and 0.0 a completely solid surface. | ||
===Quality=== | |||
To smooth your surface representation try: | To smooth your surface representation try: | ||
<source lang="python"> | <source lang="python"> | ||
| Line 19: | Line 22: | ||
or higher if you wish, though it will take longer and might look odd. | or higher if you wish, though it will take longer and might look odd. | ||
==Displaying a protein as surface with a ligand as sticks== | ==Tips== | ||
===Displaying a protein as surface with a ligand as sticks=== | |||
The easiest way to do this is to create separate objects for each type of display. | The easiest way to do this is to create separate objects for each type of display. | ||
| Line 39: | Line 43: | ||
==Calculating a partial surface== | ===Calculating a partial surface=== | ||
There is | There is, until now, an undocumented way to calculate a surface for only a part of an object without creating a new one: | ||
<source lang="python"> | <source lang="python"> | ||
flag ignore, not A/49-63/, set | flag ignore, not A/49-63/, set | ||
| Line 52: | Line 56: | ||
===Displaying surface inside a molecule=== | |||
==Displaying surface inside a molecule== | |||
As far as I can tell, setting ambient to zero alone doesn't quite do the job, since some triangles still get lit by the light source. The best combination I can find is: | As far as I can tell, setting ambient to zero alone doesn't quite do the job, since some triangles still get lit by the light source. The best combination I can find is: | ||
<source lang="python"> | <source lang="python"> | ||
| Line 70: | Line 73: | ||
</source> | </source> | ||
==Creating a Closed Surface== | ===Creating a Closed Surface=== | ||
[[Image:Surface_open.png|thumb|Example OPEN Surface|left|200px]] | [[Image:Surface_open.png|thumb|Example OPEN Surface|left|200px]] | ||
[[Image:Surface_closed.png|thumb|Example CLOSED Surface|left|200px]] | [[Image:Surface_closed.png|thumb|Example CLOSED Surface|left|200px]] | ||
Revision as of 07:33, 13 June 2005
The surface representation of a protein, in PyMol, shows the "Connolly" surface or the surface that would be traced out by the surfaces of waters in contact with teh protein at all possible positions.
Enabling
To enable the surface representation do
show surface, SEL
for any proper selection SEL.
Settings
Transparency
To adjust the transparency of surfaces try:
set transparency, 0.5
Where 1.0 will be an invisible and 0.0 a completely solid surface.
Quality
To smooth your surface representation try:
set surface_quality, 1
or higher if you wish, though it will take longer and might look odd.
Tips
Displaying a protein as surface with a ligand as sticks
The easiest way to do this is to create separate objects for each type of display.
- Load your protein - Select the ligand - Create a separate object for the ligand - Remove ligand atoms from the protein - Display both objects separately
Example:
load prot.ent,protein
select ligand,resn FAD
create lig_sticks,ligand
remove ligand
show sticks,lig_sticks
show surface,protein
Calculating a partial surface
There is, until now, an undocumented way to calculate a surface for only a part of an object without creating a new one:
flag ignore, not A/49-63/, set
delete indicate
show surface
If the surface was already computed, then you'll also need to issue the command:
rebuild
Displaying surface inside a molecule
As far as I can tell, setting ambient to zero alone doesn't quite do the job, since some triangles still get lit by the light source. The best combination I can find is:
set ambient=0
set direct=0.7
set reflect=0.0
set backface_cull=0
Which gives no shadows and only a few artifacts.
As an alternative, you might just consider showing the inside of the surface directly...that will create less visual artifacts, and so long as ambient and direct are sufficiently low, it will look reasonable in "ray".
util.ray_shadows("heavy")
set two_sided_lighting=1
set backface_cull=0
Creating a Closed Surface
To create what I'll call a closed surface (see images), you need to first make your atom selections, then create a new object for that selection then show the surface for that object. Here's an example.
# sel A, id 1-100 # create B, A # show surface, B