Annotate v: Difference between revisions
No edit summary |
No edit summary |
||
| Line 2: | Line 2: | ||
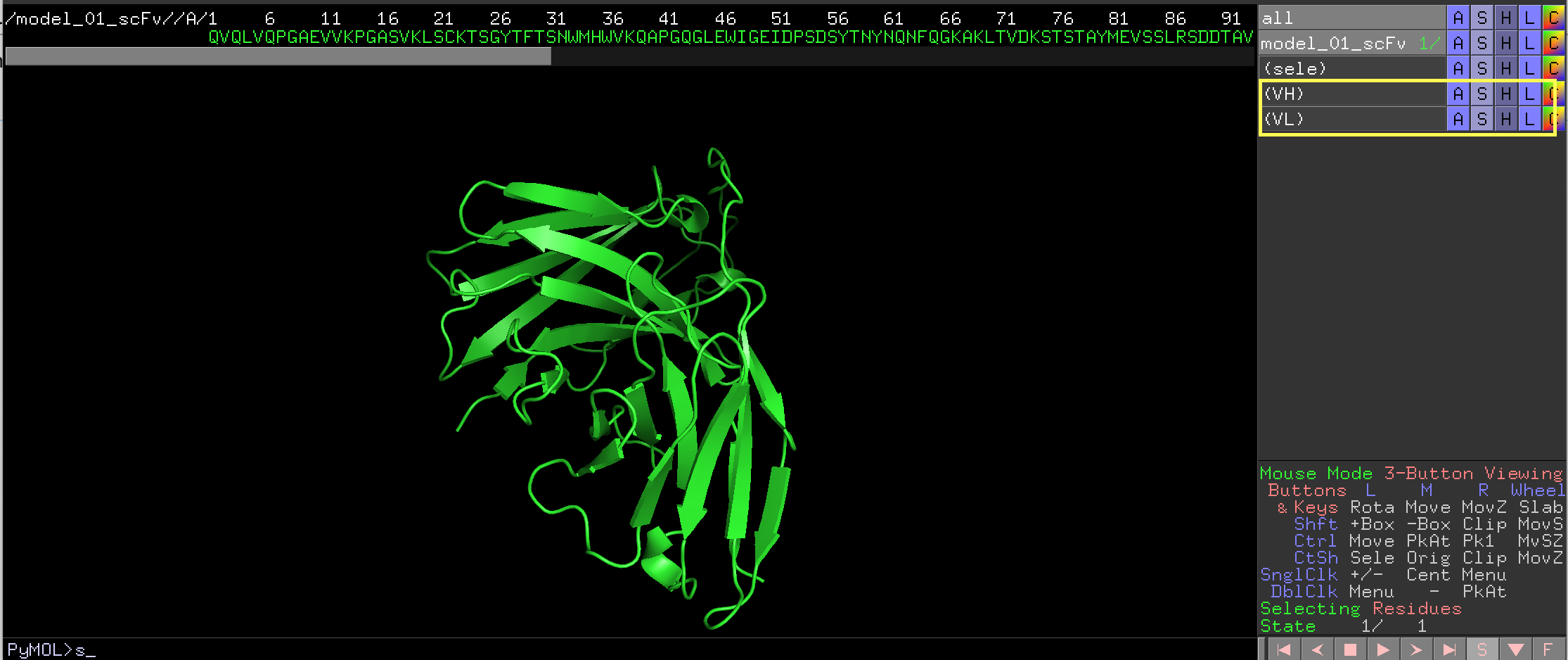
A simple-to-use script for annotation of VH or VL sequence of an antibody. It creates a Pymol object for each FR or CDR region. It utilizes the REST API interface of ''Abnum'' (http://www.bioinf.org.uk/abs/abnum/) from Dr Andrew Martin's group at UCL | A simple-to-use script for annotation of VH or VL sequence of an antibody. It creates a Pymol object for each FR or CDR region. It utilizes the REST API interface of ''Abnum'' (http://www.bioinf.org.uk/abs/abnum/) from Dr Andrew Martin's group at UCL | ||
Author: Xin Yu, 2020 | |||
== Annotation Schemes == | == Annotation Schemes == | ||
| Line 54: | Line 56: | ||
== Contact == | == Contact == | ||
Bugs? Questions? Email xinyu18018@gmail.com. Enjoy! | Bugs? Questions? Email Xin Yu @ xinyu18018@gmail.com. Enjoy! | ||
[[Category:Script_Library]] | [[Category:Script_Library]] | ||
[[Category:ObjSel_Scripts]] | [[Category:ObjSel_Scripts]] | ||
[[Category:Pymol-script-repo]] | [[Category:Pymol-script-repo]] | ||
Revision as of 16:05, 15 February 2020
Description
A simple-to-use script for annotation of VH or VL sequence of an antibody. It creates a Pymol object for each FR or CDR region. It utilizes the REST API interface of Abnum (http://www.bioinf.org.uk/abs/abnum/) from Dr Andrew Martin's group at UCL
Author: Xin Yu, 2020
Annotation Schemes
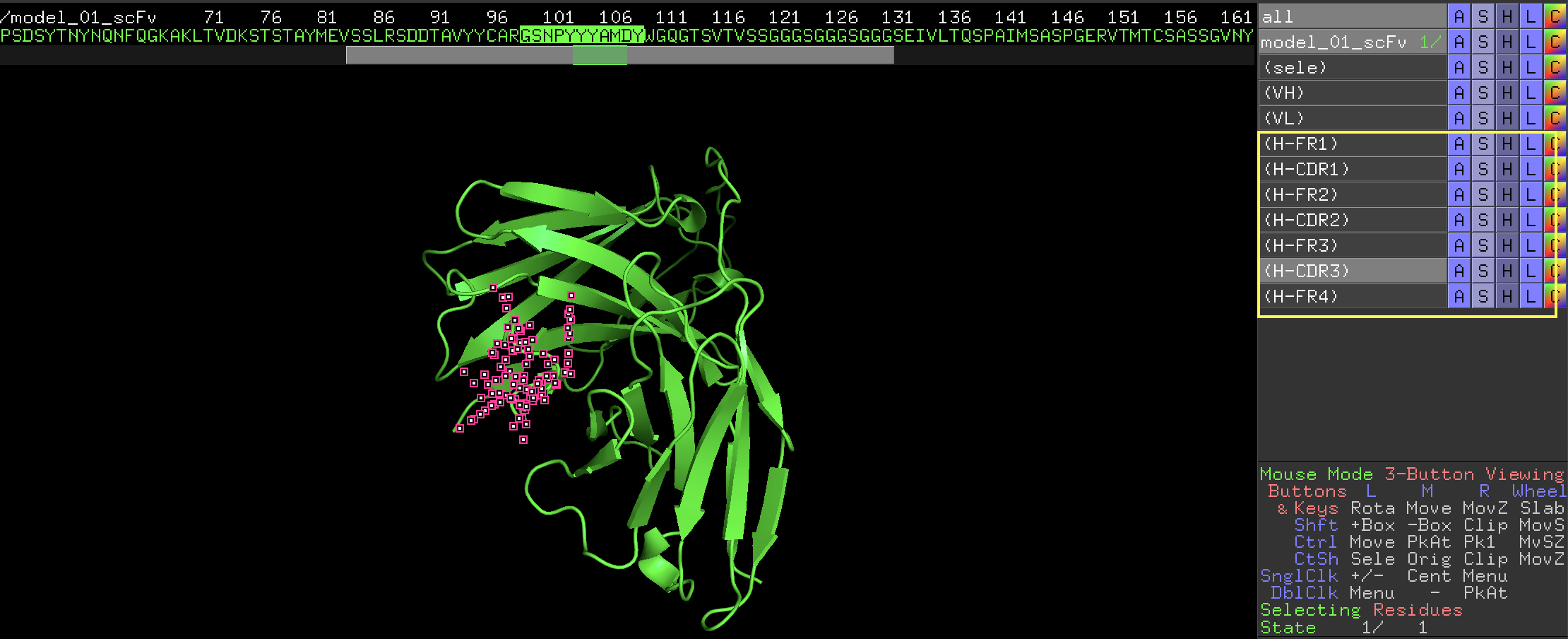
Currently supports Kabat, Chothia, Contact, IMGT1
1Definitions for Kabat, Chothia, Contact, and IIMGT are same as listed in the table (http://www.bioinf.org.uk/abs/info.html#kabatnum), except that for IMGT, H-CDR2 is defined as H51-H57 in this script, as opposed to of H51-H56 in the table. This slight revision generates result that matches that from IMGT website (http://www.imgt.org/)
Dependencies and Limitations
1. Import request module
2. Relies on internet connection to Abnum
3. Incomplete VH or VL sequence might not be annotated
How to use
1. Create a selection object for the V region, (such as VH and VL shown here). Only 1 V-region (VH OR VL) can be annotated each time. Alternatively, simply select the residues into the default <sele> group.
2. Copy the entire script and create a `.py` file. Run script.
3. The general syntax is:
annotate_v("selection_group_name", "scheme")
selection_group_name: name of the selection group where you have the input sequence. Must be single-letter amino acid coded. Either VH or VL but not both
scheme: currently supports Kabat, Chothia, Contact, IMGT. Must be lowercase
For example:
annotate_v("VH", "kabat") #input sequence from VH selection group, annotate using "kabat"
annotate_v("sele", "imgt") #input sequence from the default sele group, annotate using "imgt". You can also try "contact", "chothia"
4. In the output window, the script will print the FR and CDR regions (if found). It also automatically creates a selection group for each of the FR and CDR region.
Source code
Copy the source code into a `.py` file and run script in Pymol. Source code: https://raw.githubusercontent.com/xinyu-dev/annotate_v-for-Pymol/master/annotate_v.py
Contact
Bugs? Questions? Email Xin Yu @ xinyu18018@gmail.com. Enjoy!