Azahar: Difference between revisions
(add MCM section) |
(add change log entry) |
||
| Line 1: | Line 1: | ||
{{Infobox script-repo | {{Infobox script-repo | ||
|type = plugin | |type = plugin | ||
|download = https://github.com/agustinaarroyuelo/Azahar/ | |download = https://github.com/agustinaarroyuelo/Azahar/releases/tag/v0.8.5-beta | ||
|author = Agustina Arroyuelo and [[User:OsvaldoMartin|Osvaldo Martin]] | |author = Agustina Arroyuelo and [[User:OsvaldoMartin|Osvaldo Martin]] | ||
|license = GPL | |license = GPL | ||
| Line 60: | Line 60: | ||
== Change log == | == Change log == | ||
* 2015-10-2 (Version 0.8.5) | |||
*# Fix templates: reorder atoms, standardize nomenclature and minimize conformations | |||
*# Add new templates | |||
*# Add Monte Carlo with Minimization Routine | |||
*# fix cartoonize for 1-6 and 6-1 bonds | |||
*# add 2 new cartoon-like representations | |||
*# fix minor bugs, clean code and GUI | |||
* 2015-08-31 (Version 0.8.1) | * 2015-08-31 (Version 0.8.1) | ||
Revision as of 03:32, 2 November 2015
| Type | PyMOL Plugin |
|---|---|
| Download | https://github.com/agustinaarroyuelo/Azahar/releases/tag/v0.8.5-beta |
| Author(s) | Agustina Arroyuelo and Osvaldo Martin |
| License | GPL |
Description
Azahar (pronounced /ɑːsɑːˈɑːr/) is a plugin that extends the PyMOL's capabilities to visualize, analyze and model glycans and glycoconjugated molecules.
Installation
The plugin have been tested on Linux and Windows and should also works on Mac Os X. To install the plugin just download this zip file and install it using the plugin manager.
Azahar can be used without any dependencies (i.e. you don't need to install anything else) but if you have installed OpenBabel (and OpenBabel Python bindings) Azahar will use it to perform certain calculations like optimize the geometry of your newly created molecule. OpenBabel is also used during the Monte Carlo with Minimization routine.
If you still want to install OpenBabel on your computer see instructions here. Users of 64-bit Windows OS, please install OpenBabel 64-bit from here
Usage
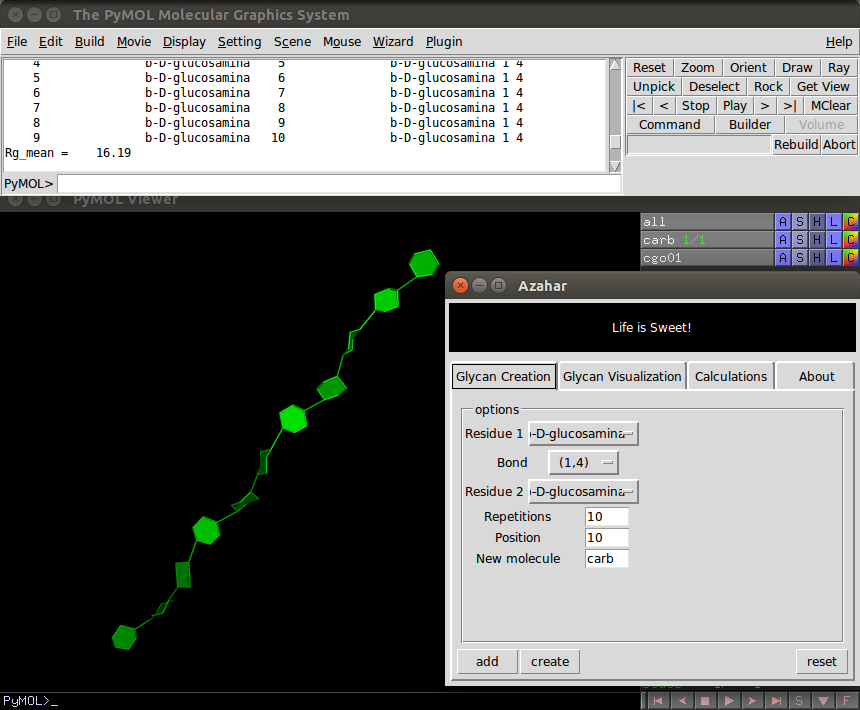
The plugin's GUI displays three interactive tabs: Creation, Visualization and Calculations. Usage is described by tab:
Glycan Creation
The Creation tab contains:
- Three option menus, two of which are used to specify residues to be connected from a list of predefined templates. And the third one, selects the chemical bond to be used.
- Three entry boxes, the first one it's called "Repetitions", where the user can enter the number of residues to be connected. The "Position" entry box indicates the index of the residue to which the next bond will be added. On a typical glycan molecule constructed using this plugin, the user would use the "Position" entry box and the bonds menu to specify ramifications. And in the "New Molecule" entry box the user can write the name of the PyMOL object corresponding to the newly created glycan molecule.
- Finally, The "add" button, when clicked, writes a text file containing the connectivity of the glycan molecule to be created. The user can edit this file or provide it as input. The connectivity instructions are also printed in PyMOL's external GUI. When the "create" button is clicked, the molecule is constructed and displayed in the PyMOL's GUI. The "reset" button resets the entry box's parameters.
Visualization
On this tab the user will find the cartoon visualization option, only available for proteins in standard PyMOL. This feature can be applied to glycan molecules created by this plugin or loaded from other sources.
Calculations
Radius of gyration and Ramachandran Plot are useful tools for the analysis of glycan molecules. Through this plugin's GUI the user can easily select a range of states to compute these calculations, and in case of the RG, the result will be displayed in the PyMOL's external GUI.
Monte Carlo
This feature will be available in the next stable release (although is already available at github).
A Monte Carlo with Minimization (MCM) is a global optimization method that have proven very efficient to sample the conformational states of complex molecules like proteins, It was first published by Li and Scheraga, and is some circles is also know as basin hopping. Briefly, the conformational search is done by perturbing a torsional angle (psi, psi, omega or chi), follow by and energy minimization and the new (minimized) conformation is accepted or rejected following the Metropolis-criteria. If the conformation is accepted the next iteration start from the new conformation, otherwise the old conformation is perturbed again.
From the GUI the user can choose the number of iterations to run, whether or not to use a SASA model to take into account the solvent contribution, and whether or not to start from a random conformation.
After the MCM procedure is completed the user will get:
- one .pdb file for each accepted conformation.
- one multi-state .pdb file (automatically uploaded at the end of the run)
- one .pdb file for the conformation with the minimum energy.
- a plain text listing the energy of each saved conformation
The internal energy is computed using the MMFF94 force field (as implemented on open-babel) and the contribution of the solvent is approximated using a SASA model (the validity of the SASA model for glycans and the relative weight of the internal and salvation energies needs further test).
Change log
- 2015-10-2 (Version 0.8.5)
- Fix templates: reorder atoms, standardize nomenclature and minimize conformations
- Add new templates
- Add Monte Carlo with Minimization Routine
- fix cartoonize for 1-6 and 6-1 bonds
- add 2 new cartoon-like representations
- fix minor bugs, clean code and GUI
- 2015-08-31 (Version 0.8.1)
- Several bugs fixed, including problems when creating branches.
- 2015-08-28 (Version 0.8)
- First beta version and first public version (several bugs expected!).