MSMS: Difference between revisions
Jump to navigation
Jump to search
Hongbo zhu (talk | contribs) No edit summary |
Hongbo zhu (talk | contribs) |
||
| Line 13: | Line 13: | ||
= External links = | = External links = | ||
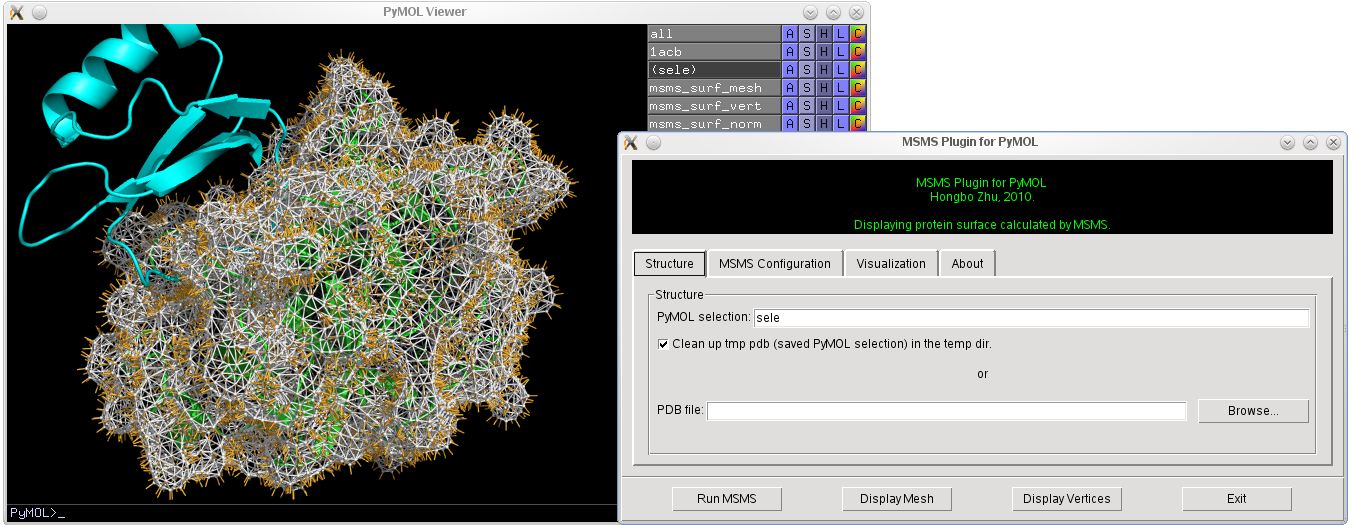
[[File:Example.png|400px|thumb|right|Demonstration of MSMS plugin.]] | [[File:Example.png|400px|thumb|right|Demonstration of MSMS plugin.]] | ||
* [http://www.netsci.org/Science/Compchem/feature14e.html Molecular Surfaces] | |||
* Most recent code on github: [https://github.com/hongbo-zhu-cn/Pymol-script-repo/blob/master/plugins/msms.py MSMS Plugin for PyMOL @ GitHub] | * Most recent code on github: [https://github.com/hongbo-zhu-cn/Pymol-script-repo/blob/master/plugins/msms.py MSMS Plugin for PyMOL @ GitHub] | ||
* Original site (not updated anymore): [http://www.biotec.tu-dresden.de/~hongboz/msms_pymol/msms_pymol.html MSMS Plugin for PyMOL] | * Original site (not updated anymore): [http://www.biotec.tu-dresden.de/~hongboz/msms_pymol/msms_pymol.html MSMS Plugin for PyMOL] | ||
Revision as of 05:05, 12 April 2012
| Type | PyMOL Plugin |
|---|---|
| Download | plugins/msms.py |
| Author(s) | Hongbo Zhu |
| License | - |
| This code has been put under version control in the project Pymol-script-repo | |
Introduction
MSMS is an excellent tool for computing protein solvent excluded surface (SES). MSMS Plugin for PyMOL provides a graphical user interface for running MSMS and displaying its results in PyMOL. Most recent MSMS Plugin code can be obtained from this link.
Only works in Linux, since pdb_to_xyzrn is a shell script. Windows users please contact author for a script replacing pdb_to_xyzrn.
External links
- Molecular Surfaces
- Most recent code on github: MSMS Plugin for PyMOL @ GitHub
- Original site (not updated anymore): MSMS Plugin for PyMOL