Cheshift: Difference between revisions
(Created page with "{{Infobox script-repo |type = plugin |filename = plugins/cheshift.py |author = Osvaldo Martin |license = GPL }} ---CheShift plug-in: Validate yo...") |
mNo edit summary |
||
| Line 2: | Line 2: | ||
|type = plugin | |type = plugin | ||
|filename = plugins/cheshift.py | |filename = plugins/cheshift.py | ||
|author = [[ | |author = [[mailto:aloctavodia@gmail.com|Osvaldo Martin]] | ||
|license = GPL | |license = GPL | ||
}} | }} | ||
| Line 21: | Line 12: | ||
chemical shifts. All computations run on CheShift server (www.cheshift.com), | chemical shifts. All computations run on CheShift server (www.cheshift.com), | ||
and the results are retrieved to PyMOL, hence an Internet connection is needed. | and the results are retrieved to PyMOL, hence an Internet connection is needed. | ||
Version : 1.0 | |||
Citation: Martin O.A. Vila J.A. and Scheraga H.A. CheShift-2: Graphic | |||
validation of protein structures. Bioinformatics 2012 (submitted) | |||
| Line 108: | Line 103: | ||
== References == | == References == | ||
Martin O.A. Vila J.A. and Scheraga H.A. (2012). CheShift-2: Graphic validation of protein structures. Bioinformatics 2012 (submitted). | |||
Vila J.A. Arnautova Y.A. Martin O.A. and Scheraga, H.A. (2009). Quantum-mechanics-derived 13C chemical shift server (CheShift) for protein structure validation. PNAS, 106(40), 16972-16977. | |||
Revision as of 02:28, 14 February 2012
| Type | PyMOL Plugin |
|---|---|
| Download | plugins/cheshift.py |
| Author(s) | [Martin] |
| License | GPL |
| This code has been put under version control in the project Pymol-script-repo | |
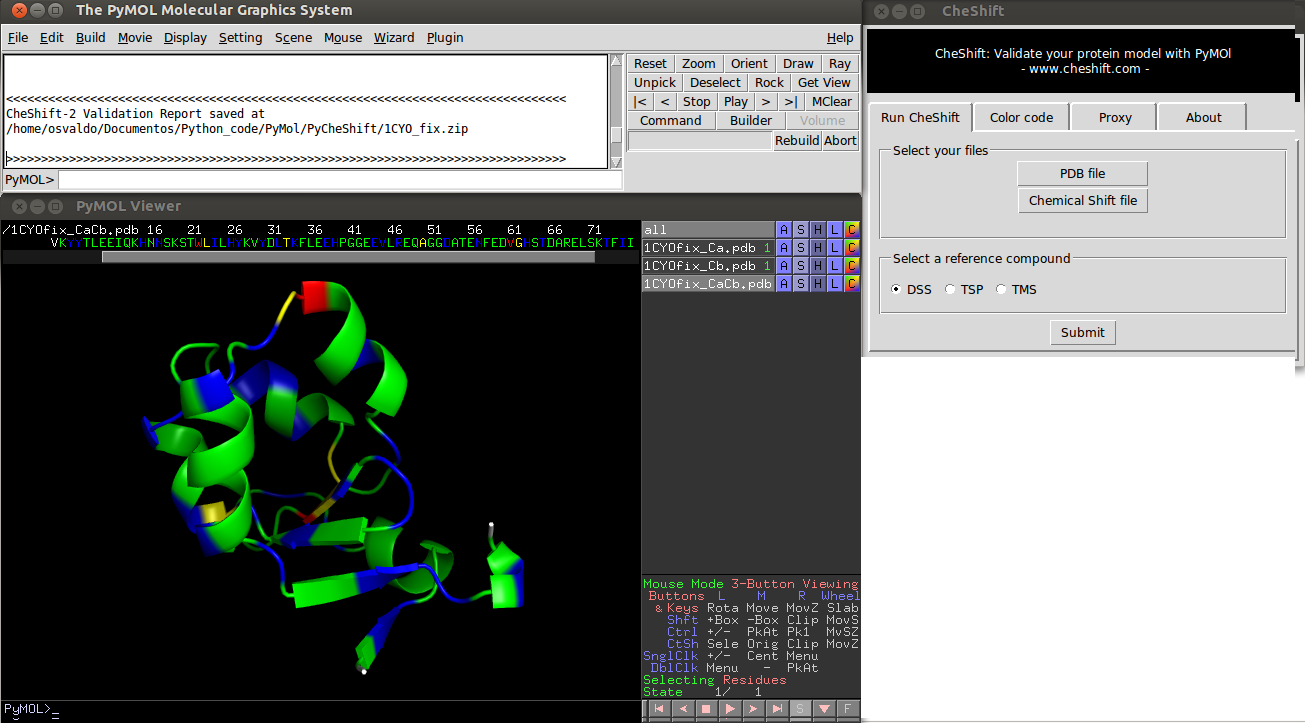
Description
This plug-in provides a way to validate a protein model using observed chemical shifts. All computations run on CheShift server (www.cheshift.com), and the results are retrieved to PyMOL, hence an Internet connection is needed.
Version : 1.0 Citation: Martin O.A. Vila J.A. and Scheraga H.A. CheShift-2: Graphic validation of protein structures. Bioinformatics 2012 (submitted)
Installation
Linux
1) The plug-in can be downloaded from here Pymol-script-repo
2) You should install mechanize, a python module. This module is available from the repositories of the main Linux distributions. Just use your default package manager (or command line) to install it. In Ubuntu/Debian the package name is "python-mechanize".
Windows
This plug-in have not been extensively tested on a Windows machines, but it should work...
1) The plug-in can be downloaded from here Pymol-script-repo
2) You should install mechanize, a python module. In order to do that, download this unzip and copy the mechanize folder were you have installed PyMol, usually is C:\Program Files\DeLano Scientific\PyMOL\modules\pmg_tk\startup
Mac OsX
This plug-in have not been tested on a Mac OsX machine, but it should work...
1) The plug-in can be downloaded from here Pymol-script-repo
2) You should install mechanize, a python module. In order to do that download this unzip and copy the mechanize folder were you have installed PyMol. For the X11/Hybrid version, the location is probably: PyMOLX11Hybrid.app/pymol/modules/pmg_tk/startup
Using the Plug-in
1) Launch PyMOL and select "CheShift" from the Plug-ins menu. 2) Select a PDB file of your protein model (using the CheShift plug-in). 3) Select a file with the experimental Chemical Shift values. 4) Click the "Submit" button. 5) Wait until results are displayed (this could take a few minutes, depending on all existing request on the server. Analyzing 20 conformations of a 100 residues protein takes ~4 minutes).
Note:
- If more than 20 conformers are uploaded only the 20 first will be analyzed
- If the PDB file has more than one chain only the first one will be analyzed
- The PDB file must not have missing residues
- Missing observed 13Cα chemical shifts are tolerated
- The observed 13Cα chemical shifts values should be in BMRB-Star format (only
the 13Cα are needed, the other nucleus could be present but are ignored)
you can use a complete BMRB file or just a file with only the experimental chemical shifts. In the latter case you can use this format:
1 21 MET N N 121.0 0.25 1
2 21 MET H H 8.36 0.01 1
3 21 MET CA C 55.9 0.20 1
4 21 MET HA H 4.39 0.01 1
5 21 MET CB C 31.8 0.20 1
6 21 MET HB2 H 2.15 0.01 2
7 21 MET HG2 H 2.54 0.01 2
8 21 MET HE H 2.09 0.01 1
9 21 MET C C 176.6 0.10 1
10 22 GLY N N 111.4 0.25 1
11 22 GLY H H 8.55 0.01 1
12 22 GLY CA C 44.9 0.20 1
13 22 GLY HA2 H 4.15 0.01 2
14 22 GLY HA3 H 3.91 0.01 2
15 22 GLY C C 173.2 0.10 1
or this format:
1 1 1 SER HA H 4.44 0.005 1
2 1 1 SER HB2 H 3.85 0.005 2
3 1 1 SER HB3 H 3.88 0.005 2
4 1 1 SER C C 168.6 0.2 1
5 1 1 SER CA C 58.4 0.2 1
6 1 1 SER CB C 63.8 0.2 1
7 2 2 ALA H H 8.23 0.005 1
8 2 2 ALA HA H 4.87 0.005 1
9 2 2 ALA HB H 1.37 0.005 1
10 2 2 ALA C C 174.5 0.2 1
11 2 2 ALA CA C 51.7 0.2 1
12 2 2 ALA CB C 23.3 0.2 1
13 2 2 ALA N N 120.2 0.2 1
Proxy Configuration
The CheShift plug-in will try to correctly guess your proxy configuration, but this is a tricky business and many things could fail. In that scenario you will be prompted to manually set your proxy settings. In case you manually set the proxy configuration the plug-in will save your proxy settings and next time it will attempt to use those saved settings to connect to the Internet.
License
CheShift plug-in is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License. A complete copy of the GNU General Public License can be accessed here http://www.gnu.org/licenses/.
CheShift Server (www.cheshift.com) is available free of charge ONLY for academic use.
References
Martin O.A. Vila J.A. and Scheraga H.A. (2012). CheShift-2: Graphic validation of protein structures. Bioinformatics 2012 (submitted). Vila J.A. Arnautova Y.A. Martin O.A. and Scheraga, H.A. (2009). Quantum-mechanics-derived 13C chemical shift server (CheShift) for protein structure validation. PNAS, 106(40), 16972-16977.