Pseudoatom: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
(ARGUMENTS) |
||
| (One intermediate revision by one other user not shown) | |||
| Line 1: | Line 1: | ||
[[pseudoatom]] creates a molecular object with a pseudoatom or adds a pseudoatom to a molecular object if the specified object already exists. Default position is in the middle of the viewing window. | |||
== USAGE == | |||
<source lang="python"> | <source lang="python"> | ||
pseudoatom object [, selection [, name [, resn [, resi [, chain | pseudoatom object [, selection [, name [, resn [, resi [, chain | ||
| Line 23: | Line 8: | ||
[, pos [, state [, mode [, quiet ]]]]]]]]]]]]]]]]] | [, pos [, state [, mode [, quiet ]]]]]]]]]]]]]]]]] | ||
</source> | </source> | ||
== | == ARGUMENTS == | ||
* object = string: name of object to create or modify | |||
* selection = string: optional atom selection. If given, calculate position (pos) as center of this selection (like [[Center Of Mass|center of mass]] without atom masses). | |||
* name, resn, resi, chain, segi, elem, vdw, hetatm, b, q = string: [[Property_Selectors|atom properties]] of pseutoatom | |||
* color = string: [[color]] of pseudoatom | |||
* label = string: place a [[label|text label]] and hide all other representations | |||
* pos = 3-element tuple of floats: location in space (only if no selection given) | |||
* state = integer: [[state]] to modify, 0 for current state, -1 for all states {default: 0} | |||
* mode = string: determines the vdw property if vdw is not given. Either as RMS distance (mode=rms, like [[radius of gyration]] without atom masses) or as maximum distance (mode=extent) from pseudoatom to selection {default: rms} | |||
<source lang="python"> | <source lang="python"> | ||
# create the pseudoatom | # create the pseudoatom | ||
pseudoatom tmpPoint | pseudoatom tmpPoint | ||
ObjMol: created tmpPoint/PSDO/P/PSD`1/PS1 | |||
# show it as a sphere. | # show it as a sphere. | ||
show spheres, tmpPoint | show spheres, tmpPoint | ||
| Line 35: | Line 36: | ||
# create another, with more options. | # create another, with more options. | ||
pseudoatom tmpPoint2, resi=40, chain=ZZ, b=40, color=tv_blue, pos=[-10, 0, 10] | pseudoatom tmpPoint2, resi=40, chain=ZZ, b=40, color=tv_blue, pos=[-10, 0, 10] | ||
ObjMol: created tmpPoint2/PSDO/ZZ/PSD`40/PS1 | |||
</source> | |||
== EXAMPLES FOR USE == | |||
[[pseudoatom]] can be used for a wide variety of tasks where on must place an atom or a label in 3D space, e.g. as a placeholder for distance measurement or distance specifications. | |||
# | <source lang="python"> | ||
# A pseudoatom as a placeholder for selections according to distance: | |||
load $TUT/1hpv.pdb | load $TUT/1hpv.pdb | ||
pseudoatom tmp, pos=[10.0, 17.0, -3.0] | pseudoatom tmp, pos=[10.0, 17.0, -3.0] | ||
| Line 42: | Line 50: | ||
delete tmp | delete tmp | ||
# | # A pseudoatom as placeholder for distance measurement: | ||
# position it at the center of an aromatic ring. Then | |||
# calc the distance from another atom to the pseudoatom. | |||
load $TUT/1hpv.pdb | load $TUT/1hpv.pdb | ||
pseudoatom pi_cent,b/53/cg+cz | pseudoatom pi_cent,b/53/cg+cz | ||
| Line 50: | Line 58: | ||
</source> | </source> | ||
== | You can use a [[pseudoatom]] to make a label for a scene title. Move the protein to the bottom of the window before the pseudoatom is created. Or move the label after creating it (Shift + Middle mouse button in editing mode). | ||
<source lang="python"> | |||
fetch 1rq5 | |||
pseudoatom forLabel | |||
label forLabel, "This Protein is a Carbohydrate Active Enzyme" | |||
set label_color, black | |||
# png ray=1 | |||
</source> | |||
<gallery widths=300px heights=300px> | |||
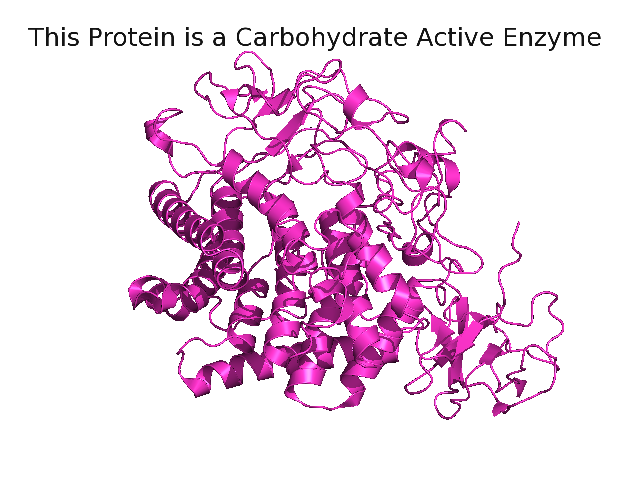
Image:Patom.png|You can use a pseudoatom for a movable scene label. | |||
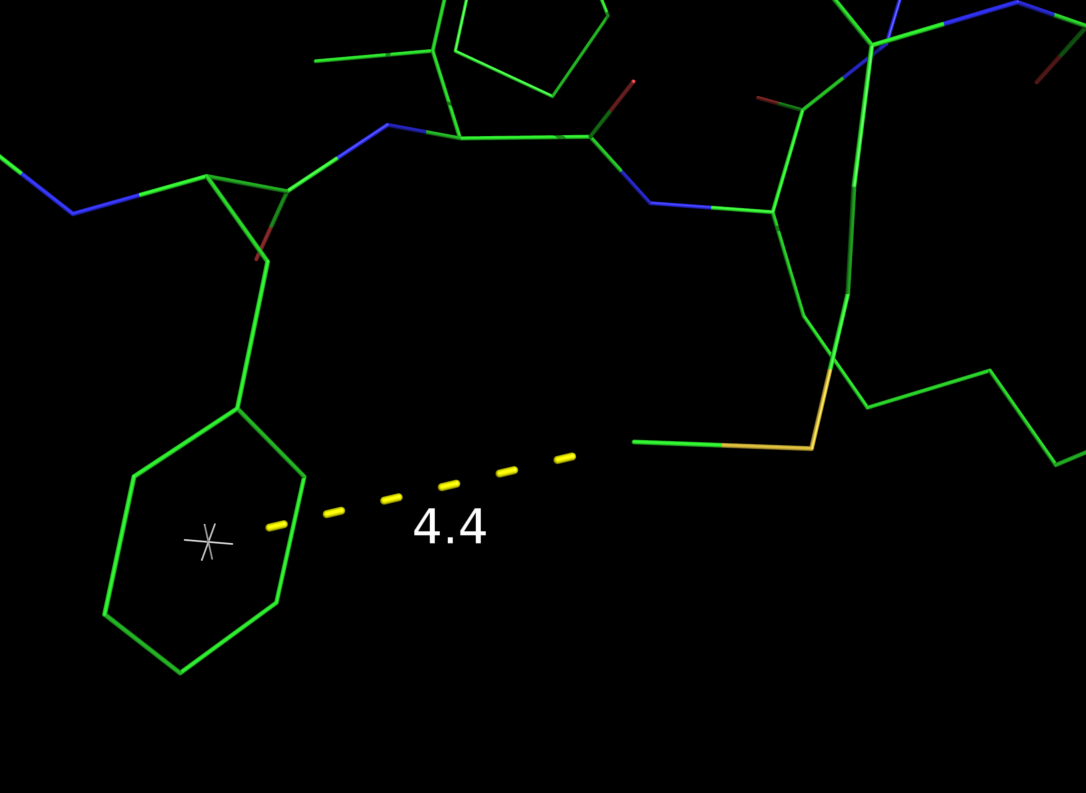
Image:Pseu1.png|Example of using a pseudoatom for distance measurement. | |||
</gallery> | |||
== References == | == References == | ||
Latest revision as of 08:40, 24 May 2011
pseudoatom creates a molecular object with a pseudoatom or adds a pseudoatom to a molecular object if the specified object already exists. Default position is in the middle of the viewing window.
USAGE
pseudoatom object [, selection [, name [, resn [, resi [, chain
[, segi [, elem [, vdw [, hetatm [, b [, q [, color [, label
[, pos [, state [, mode [, quiet ]]]]]]]]]]]]]]]]]
ARGUMENTS
- object = string: name of object to create or modify
- selection = string: optional atom selection. If given, calculate position (pos) as center of this selection (like center of mass without atom masses).
- name, resn, resi, chain, segi, elem, vdw, hetatm, b, q = string: atom properties of pseutoatom
- color = string: color of pseudoatom
- label = string: place a text label and hide all other representations
- pos = 3-element tuple of floats: location in space (only if no selection given)
- state = integer: state to modify, 0 for current state, -1 for all states {default: 0}
- mode = string: determines the vdw property if vdw is not given. Either as RMS distance (mode=rms, like radius of gyration without atom masses) or as maximum distance (mode=extent) from pseudoatom to selection {default: rms}
# create the pseudoatom
pseudoatom tmpPoint
ObjMol: created tmpPoint/PSDO/P/PSD`1/PS1
# show it as a sphere.
show spheres, tmpPoint
# create another, with more options.
pseudoatom tmpPoint2, resi=40, chain=ZZ, b=40, color=tv_blue, pos=[-10, 0, 10]
ObjMol: created tmpPoint2/PSDO/ZZ/PSD`40/PS1
EXAMPLES FOR USE
pseudoatom can be used for a wide variety of tasks where on must place an atom or a label in 3D space, e.g. as a placeholder for distance measurement or distance specifications.
# A pseudoatom as a placeholder for selections according to distance:
load $TUT/1hpv.pdb
pseudoatom tmp, pos=[10.0, 17.0, -3.0]
show sticks, tmp expand 6
delete tmp
# A pseudoatom as placeholder for distance measurement:
# position it at the center of an aromatic ring. Then
# calc the distance from another atom to the pseudoatom.
load $TUT/1hpv.pdb
pseudoatom pi_cent,b/53/cg+cz
dist pi_cent////ps1, b/met`46/ce
You can use a pseudoatom to make a label for a scene title. Move the protein to the bottom of the window before the pseudoatom is created. Or move the label after creating it (Shift + Middle mouse button in editing mode).
fetch 1rq5
pseudoatom forLabel
label forLabel, "This Protein is a Carbohydrate Active Enzyme"
set label_color, black
# png ray=1
References
PyMOL Mailing List