All states: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| (One intermediate revision by the same user not shown) | |||
| Line 11: | Line 11: | ||
==Example== | ==Example== | ||
<source lang="python"> | <source lang="python"> | ||
# fetch a PDB and show it in multiple states; this one | |||
# line does the work of the next 6 lines. | |||
fetch 1nmr | |||
# this is older code, use the above code which is newer | |||
import urllib2 | import urllib2 | ||
pdbCode = '1BRV' | pdbCode = '1BRV' | ||
| Line 34: | Line 39: | ||
[[Category:Settings|All states]] | [[Category:Settings|All states]] | ||
[[Category:States]] | |||
Latest revision as of 08:54, 19 January 2009
Overview
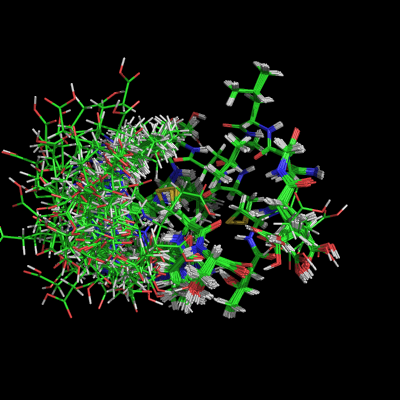
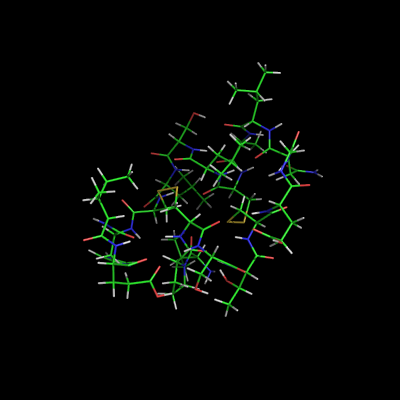
When set "on", this setting causes PyMOL to display all states or in NMR jargon: all the models in the ensemble. The 'default' behavior (OFF) can be overridden by placing the "set all_states, on" statement into your '.pymolrc' file, located in your login directory (under all flavors of unix).
Syntax
set all_states, on
set all_states, off
Example
# fetch a PDB and show it in multiple states; this one
# line does the work of the next 6 lines.
fetch 1nmr
# this is older code, use the above code which is newer
import urllib2
pdbCode = '1BRV'
pdbUrl = 'http://www.rcsb.org/pdb/downloadFile.do?fileFormat=pdb&compression=NO&structureId='+pdbCode
pdbFile = urllib2.urlopen(pdbUrl)
pdbContent = pdbFile.read()
cmd.read_pdbstr(pdbContent, pdbCode)
set all_states, on
This shows the effect of turning on/off the all_states setting used with the script above.
User Notes
- There was an error with importing ensembles of ensembles files (complex MOL2 files) before revision 3541. If you experience this problem, update to 3541 or later.