Dockingpie: Difference between revisions
(DockingPie: a Consensus Docking plugin for PyMOL) |
No edit summary |
||
| (5 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
== Description == | == Description == | ||
'''DockingPie''' | '''DockingPie''': a Molecular Docking plugin for PyMOL [1]. | ||
[[Tutorial:Plugins:Threads]] | |||
DockingPie implements: '''Smina''', '''Autodock Vina''', '''RxDock''' and '''ADFR''' [2-5]. | |||
Providing an easy interface to four docking programs, DockingPie is particularly suited as a platform to carry out consensus docking and scoring analyses. | Providing an easy interface to four docking programs, DockingPie is particularly suited as a platform to carry out consensus docking and scoring analyses. | ||
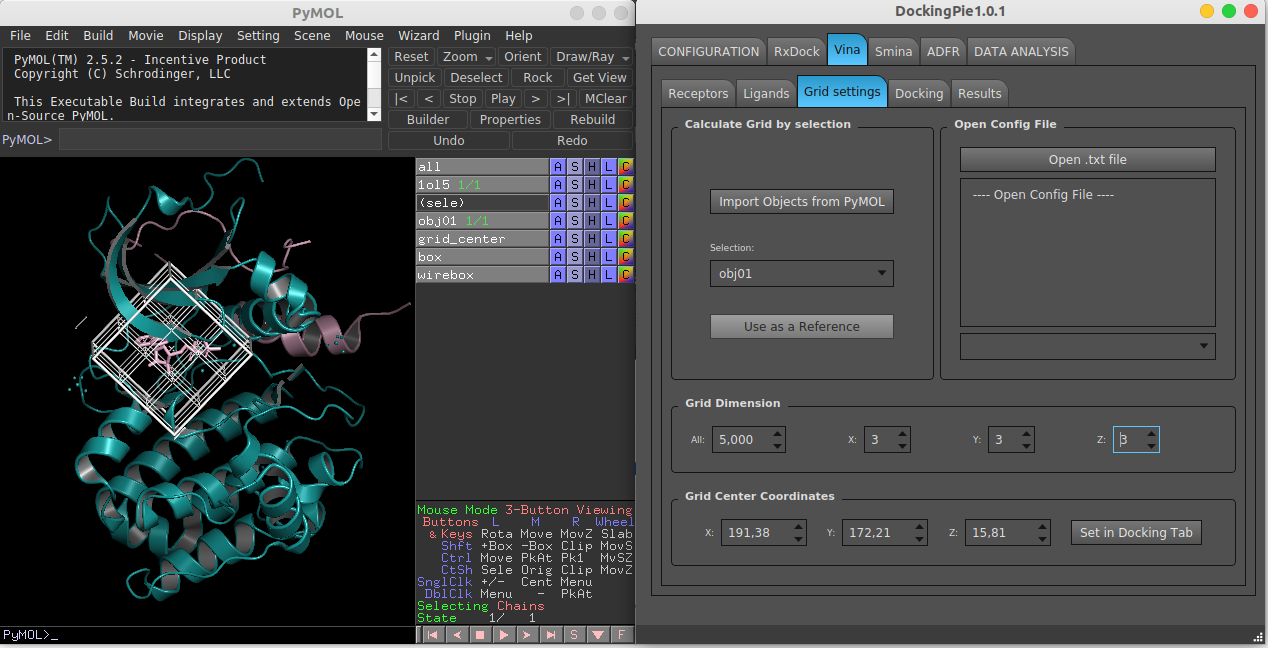
[[File:dockingpie.png|DockingPie]] | [[File:dockingpie.png|600px|thumb|center|DockingPie]] | ||
== Requirements == | == Requirements == | ||
| Line 32: | Line 32: | ||
A detailed explanation on how to use DockingPie, some videos and tutorials can be found at the following links. | A detailed explanation on how to use DockingPie, some videos and tutorials can be found at the following links. | ||
User's Guide: [https://github.com/paiardin/DockingPie/releases/download/versioning/DockingPie_User_Guide.pdf download from here] | * User's Guide: [https://github.com/paiardin/DockingPie/releases/download/versioning/DockingPie_User_Guide.pdf download from here] | ||
* DockingPie GitHub Wiki: [https://github.com/paiardin/DockingPie/wiki Home] [https://github.com/paiardin/DockingPie/wiki/Tutorials Tutorials] | |||
* Website: [http://schubert.bio.uniroma1.it Structural Bioinformatics Group at Sapienza] | |||
== Configuration == | |||
DockingPie, at the current and first release, integrates four different docking programs: RxDock [2], Vina [3], Smina [4] and ADFR [5]; several chemo-informatics python modules (i.e. AutoDockTools [6], Openbabel [7], sPyRMSD [8]) and other external tools like sdsorter [9]. The CONFIGURATION tab provides an easy way for the installation of the needed tools from within the plugin in two steps, as reported next. | |||
* Configure external tools: CONFIGURATION tab → Configure → Start Download → Finish Download | |||
* Install external tools: If the needed tools are not currently installed on the user’s machine, the *Install* button is enabled and it can be used to install the external components. | |||
== How to Cite DockingPie == | == How to Cite DockingPie == | ||
Latest revision as of 07:29, 19 July 2023
Description
DockingPie: a Molecular Docking plugin for PyMOL [1]. Tutorial:Plugins:Threads DockingPie implements: Smina, Autodock Vina, RxDock and ADFR [2-5].
Providing an easy interface to four docking programs, DockingPie is particularly suited as a platform to carry out consensus docking and scoring analyses.
Requirements
Minimal requirement: a recent version of PyMOL installed on your computer.
DockingPie is compatible with incentive PyMOL builds distributed by Schrodinger (required PyMOL version >= 2.3.4) and open source builds (required PyMOL version >= 2.3.0).
DockingPie is distributed freely to the public and it has been tested and runs on Windows, macOS and Linux versions of PyMOL.
(Some incompatibilities may arise with the usage of PyMOL version 2.5.x if ‘undo’ function is enabled, which in PyMOL 2.5.2 still shows some shortcomings. Therefore, when the plugin is opened, the ‘undo’ function is automatically disabled and it is strongly suggested to keep it disabled when using the plugin.)
DockingPie Download
Download: https://github.com/paiardin/DockingPie/archive/refs/heads/main.zip
DockingPie Install
DockingPie is installed, as any other PyMOL [1] plugin, via the PyMOL plugin manager:
First download the latest version of the plugin ZIP file here
Launch PyMOL and use the Plugin → Plugin Manager command from the main menu of PyMOL. The plugin manager window of PyMOL will open.
Click on Install New Plugin and press the Choose File… button. Select the DockingPie ZIP file which you have downloaded before. You will be asked to give the path of the directory in which to install the plugin files. Just select the default option if you are unsure about what to do (the location of the plugin files does not make any difference when running the plugin).
How to use DockingPie
A detailed explanation on how to use DockingPie, some videos and tutorials can be found at the following links.
- User's Guide: download from here
Configuration
DockingPie, at the current and first release, integrates four different docking programs: RxDock [2], Vina [3], Smina [4] and ADFR [5]; several chemo-informatics python modules (i.e. AutoDockTools [6], Openbabel [7], sPyRMSD [8]) and other external tools like sdsorter [9]. The CONFIGURATION tab provides an easy way for the installation of the needed tools from within the plugin in two steps, as reported next.
- Configure external tools: CONFIGURATION tab → Configure → Start Download → Finish Download
- Install external tools: If the needed tools are not currently installed on the user’s machine, the *Install* button is enabled and it can be used to install the external components.
How to Cite DockingPie
If you find DockingPie useful, please cite:
Serena Rosignoli and Alessandro Paiardini, DockingPie: a consensus docking plugin for PyMOL, Bioinformatics, 2022, btac452, DOI
References
[1] DeLano, WL. (2002). “The PyMOL Molecular Graphics System on World Wide Web.” CCP4 Newsletter On Protein Crystallography
[2] Ruiz-Carmona, S., Alvarez-Garcia, D., Foloppe, N., Garmendia-Doval, A. B., Juhos, S., Schmidtke, P., Barril, X., Hubbard, R. E., & Morley, S. D. (2014). rDock: a fast, versatile and open source program for docking ligands to proteins and nucleic acids. PLoS computational biology, 10(4), e1003571. https://doi.org/10.1371/journal.pcbi.1003571
[3] O. Trott, A. J. Olson, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading, Journal of Computational Chemistry 31 (2010) 455-461
[4] Koes, David Ryan et al. “Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise.”Journal of chemical information and modeling vol. 53,8 (2013): 1893-904. doi:10.1021/ci300604z
[5] Ravindranath PA, Forli S, Goodsell DS, Olson AJ, Sanner MF. AutoDockFR: Advances in Protein-Ligand Docking with Explicitly Specified Binding Site Flexibility. PLoS Comput Biol. 2015;11(12):e1004586. Published 2015 Dec 2. doi:10.1371/journal.pcbi.1004586
[6] Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791.
[7] O'Boyle, N.M., Banck, M., James, C.A. et al. Open Babel: An open chemical toolbox. J Cheminform 3, 33 (2011). https://doi.org/10.1186/1758-2946-3-33
[8] Meli, R., Biggin, P.C. spyrmsd: symmetry-corrected RMSD calculations in Python. J Cheminform 12, 49 (2020). https://doi.org/10.1186/s13321-020-00455-2
[9] https://sourceforge.net/projects/sdsorter/
[10] Anaconda Software Distribution. (2020). Anaconda Documentation. Anaconda Inc. Retrieved from https://docs.anaconda.com/