Bnitools: Difference between revisions
Jump to navigation
Jump to search
mNo edit summary |
No edit summary |
||
| (9 intermediate revisions by 2 users not shown) | |||
| Line 5: | Line 5: | ||
|license = BSD | |license = BSD | ||
}} | }} | ||
[[File:bnitools.png||thumb|right|Screenshot of the BNI Tools plugin menu]] | |||
'''BNI Tools''' is a plug in for PyMOL which adds additional functionalities and presets to the PyMOL GUI. | |||
== Installation == | |||
Install it using the 'Install Plugin' menu within PyMOL:<br> | |||
'''PyMOL > Plugin > Plugin Manager'''<br> | |||
== Example images created using BNI Tools == | |||
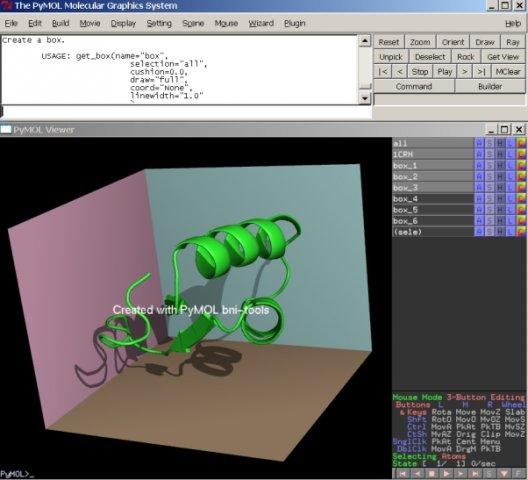
[[File:bni_box.jpg|150px|||Create a box around a selection]] | |||
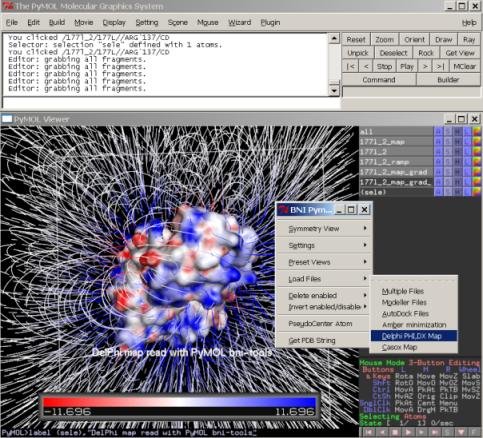
[[File:bni_delphimap.jpg|150px|||Load and show DelPhi or other maps]] | |||
[[File:bni_box2.jpg|150px|||Create a box]] | |||
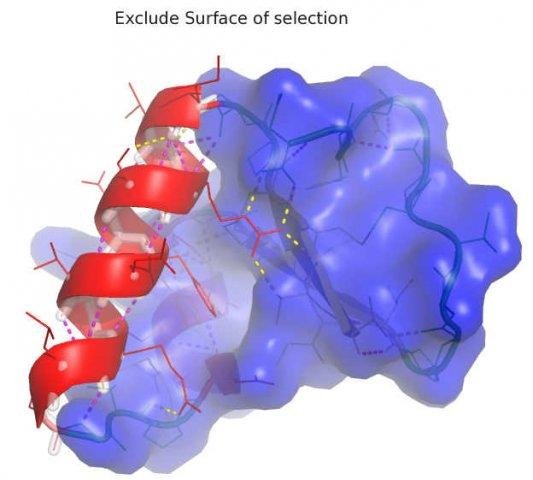
[[File:Bni_excludesurf.jpg|150px|||Exclude parts of surface]] | |||
[[File:Bni_plane.jpg|150px|||Create Plane]] | |||
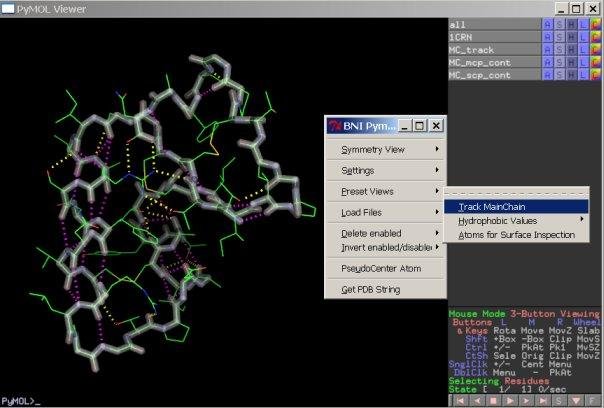
[[File:Bni_track.jpg|150px|||Create a main chain track]] | |||
== BNI Tools dropdown menu (Plugin > BNI Tools > ...) == | |||
[+]Load Files-> | |||
[-]modeller files | |||
* load multiple modeller files and sort by | |||
* objective function energy | |||
* and add energy to object title | |||
[-]autodock files | |||
* load autodock .dlg or .dlg.pdb file into | |||
* different states with energy in object | |||
* title | |||
[-]amber minimization | |||
* load amber minimization file with energy in | |||
* title if the .info file is present in same | |||
* directory and has the same name as the pdb file. | |||
[-]delphi phi,dx map | |||
* load delphi map and corresponding pdb file | |||
* simultaneously and show surface colored by | |||
* PHI or DX map. (show the surface to see the | |||
* effect) | |||
[-]casox map | |||
* load casox map (cavity calculation) and | |||
* show ligsite accessibility | |||
* value maps. (7 closed cavity to 1 open) | |||
[-]multiple files into states | |||
* load multiple pdb files (e.g. MD simulation | |||
* snapshots) into one state. (object is named | |||
* by first object loaded) | |||
[+]Fetch-> | |||
[-]Density View (EDS) | |||
* load density and pdb file from EDS | |||
* (Electron Density Server) | |||
* if available, and show density with density | |||
* wizard | |||
[-]RCSB Biol. Assembly | |||
* load biological assembly from RCSB | |||
* protein database | |||
[-]2FoFc map(s) | |||
* load (multiple) 2FoFc maps | |||
* from EDS density server | |||
* if available | |||
[-]FoFc maps(s) | |||
* load (multiple) FoFc maps | |||
* from EDS density server | |||
* if available | |||
[+]Edit-> | |||
[-]HIS --> HID,HIE,HIP | |||
* change histidine residues to HID,HIE,HIP | |||
* depending on hydrogens on histidine | |||
[-]HID,HIE,HIP --> HIS | |||
* change altered histidine residues | |||
* back to HIS | |||
[-]Poly-Alanine Chain | |||
* create a poly alanine chain (GLY and ALA) | |||
* for molecular replacement | |||
[-]MSE --> MET | |||
* change selenomethionine to methionine | |||
[-]del alternates | |||
* delete alternates in selection | |||
[-]Unbond- > | |||
* unbond atoms in selection | |||
[+]Images-> | |||
* create ray traced images depending on | |||
* x size and resolution (dpi) | |||
[+]Create-> | |||
* create compiled graphics objects (CGO) | |||
* these objects can be altered in color or | |||
* transparency, and they can be dragged | |||
* and rotated in space by the | |||
* "action->drag" command and using | |||
* "shift" and mouse buttons. | |||
[-]Plane | |||
* create a plane (with certain cushion) | |||
* using a selection of three atoms | |||
[-]Box | |||
* create a box around selection | |||
* the whole box can be altered as group | |||
* or by side planes separately | |||
[-]Triangle | |||
* create a triangle using a selection of three | |||
* atoms | |||
[+]pseudo center atom | |||
* create pseudo atom at the center of atoms | |||
* in selection | |||
* move atoms with editing->"shift" | |||
* and middle mouse button | |||
== BNI tools integrated in PyMOL sidebar == | |||
[+]Action on "all" | |||
[-]delete enabled | |||
* delete all enabled objects or | |||
* selections | |||
[-]invert enabled/disabled | |||
* disable currently enabled objects | |||
[-]combine selections | |||
* combine all enabled or disabled | |||
* selections to selection (sele) | |||
[+]action | |||
[-]sequence | |||
* show sequence in different formats | |||
* of selection | |||
* copy and paste to text file for later use | |||
[.]fasta | |||
* show sequence of selection | |||
* in fasta format | |||
[.]pir | |||
* show sequence of selection | |||
* in pir format | |||
[.]modeller | |||
* show sequence of selection | |||
* in modeller pir format | |||
[.]list | |||
* create residue or atom lists | |||
* of selection | |||
[+]preset | |||
[-]track main chain | |||
* create a new object which tracks the | |||
* main chain atoms and shows main chain | |||
* and side chain polar contacts | |||
[-]symmetry surface | |||
* create a symmetry view of the selected | |||
* atoms showing the contact surface as well | |||
* as a selection entry of the atoms in | |||
* contact with symmetry mates | |||
* (only includes atoms of the initial selection). | |||
[-]hydrophobic residues | |||
* show hydrophobic residues | |||
* depending on hydrophobic residue scales | |||
* by KandD (Kyte & Doolittle | |||
* J Mol Biol 157:105, 1982) | |||
* Rose (Rose et al. | |||
* Science, 229, 834-838,1985) | |||
* GES (Engelman Engelman et al. | |||
* Annu Rev Biophys Biophys Chem, | |||
* 15, 321-353(1986) | |||
* (no window selection; just raw categories | |||
* are colored by: blue-hydrophile | |||
* green-neutral | |||
* red-hydrophobe ) | |||
[-]surface inspection | |||
* create selections for surface | |||
* inspections | |||
# BNI tools additional settings in PyMOL sidebar | |||
[+]color | |||
[-]by ss | |||
* color helix,sheet and loop separately | |||
[.]Helix | |||
[.]Sheet | |||
[.]Loop | |||
# this section is replaced by "[-] by_rep" in PyMOL versions >1.6 | |||
[-]surface | |||
* color surface separately from atoms | |||
[.]by atom | |||
* set surface color to standard | |||
[.]by map | |||
* if a map or ramp is loaded | |||
* color surface by ramp/map | |||
[.]my color | |||
* color surface by own defined color | |||
[-]mesh | |||
* color mesh separately from atoms | |||
[.]by atom | |||
* set mesh color to standard | |||
[.]by map | |||
* if a map or ramp is loaded | |||
* color mesh by ramp/map | |||
[.]my color | |||
* color mesh by own defined color | |||
[-]label | |||
* color labels separately | |||
[.]by atom | |||
* set label color by atom | |||
[.]my color | |||
* color labels by own defined color | |||
[-]stick | |||
* color sticks separately | |||
[.]standard | |||
* set stick color to standard | |||
[.]my color | |||
* color sticks by own defined color | |||
[-]my colors | |||
* use/append own defined colors | |||
* own colors can be defined by | |||
* Setting->Colors..->New | |||
* to keep color settings for | |||
* other pymol sessions | |||
* you have to set the colors | |||
* in .pymolrc or similar | |||
* pymol setting file | |||
* like | |||
* set_color mycolor,[ 1.00, 1.00, 1.00] | |||
[+]show | |||
[-]surface flag | |||
* set surface flag of atoms to show hide | |||
* or ignore fro surface calculation | |||
[-]transparency | |||
* set different transparency types | |||
* on selections or atoms | |||
[[Category:Plugins]] | |||
[[Category:Pymol-script-repo]] | |||
Latest revision as of 09:45, 12 May 2016
| Type | PyMOL Plugin |
|---|---|
| Download | plugins/bnitools.py |
| Author(s) | Georg Steinkellner |
| License | BSD |
| This code has been put under version control in the project Pymol-script-repo | |
BNI Tools is a plug in for PyMOL which adds additional functionalities and presets to the PyMOL GUI.
Installation
Install it using the 'Install Plugin' menu within PyMOL:
PyMOL > Plugin > Plugin Manager
Example images created using BNI Tools
[+]Load Files->
[-]modeller files
* load multiple modeller files and sort by
* objective function energy
* and add energy to object title
[-]autodock files
* load autodock .dlg or .dlg.pdb file into
* different states with energy in object
* title
[-]amber minimization
* load amber minimization file with energy in
* title if the .info file is present in same
* directory and has the same name as the pdb file.
[-]delphi phi,dx map
* load delphi map and corresponding pdb file
* simultaneously and show surface colored by
* PHI or DX map. (show the surface to see the
* effect)
[-]casox map
* load casox map (cavity calculation) and
* show ligsite accessibility
* value maps. (7 closed cavity to 1 open)
[-]multiple files into states
* load multiple pdb files (e.g. MD simulation
* snapshots) into one state. (object is named
* by first object loaded)
[+]Fetch->
[-]Density View (EDS)
* load density and pdb file from EDS
* (Electron Density Server)
* if available, and show density with density
* wizard
[-]RCSB Biol. Assembly
* load biological assembly from RCSB
* protein database
[-]2FoFc map(s)
* load (multiple) 2FoFc maps
* from EDS density server
* if available
[-]FoFc maps(s)
* load (multiple) FoFc maps
* from EDS density server
* if available
[+]Edit->
[-]HIS --> HID,HIE,HIP
* change histidine residues to HID,HIE,HIP
* depending on hydrogens on histidine
[-]HID,HIE,HIP --> HIS
* change altered histidine residues
* back to HIS
[-]Poly-Alanine Chain
* create a poly alanine chain (GLY and ALA)
* for molecular replacement
[-]MSE --> MET
* change selenomethionine to methionine
[-]del alternates
* delete alternates in selection
[-]Unbond- >
* unbond atoms in selection
[+]Images->
* create ray traced images depending on
* x size and resolution (dpi)
[+]Create->
* create compiled graphics objects (CGO)
* these objects can be altered in color or
* transparency, and they can be dragged
* and rotated in space by the
* "action->drag" command and using
* "shift" and mouse buttons.
[-]Plane
* create a plane (with certain cushion)
* using a selection of three atoms
[-]Box
* create a box around selection
* the whole box can be altered as group
* or by side planes separately
[-]Triangle
* create a triangle using a selection of three
* atoms
[+]pseudo center atom
* create pseudo atom at the center of atoms
* in selection
* move atoms with editing->"shift"
* and middle mouse button
BNI tools integrated in PyMOL sidebar
[+]Action on "all"
[-]delete enabled
* delete all enabled objects or
* selections
[-]invert enabled/disabled
* disable currently enabled objects
[-]combine selections
* combine all enabled or disabled
* selections to selection (sele)
[+]action
[-]sequence
* show sequence in different formats
* of selection
* copy and paste to text file for later use
[.]fasta
* show sequence of selection
* in fasta format
[.]pir
* show sequence of selection
* in pir format
[.]modeller
* show sequence of selection
* in modeller pir format
[.]list
* create residue or atom lists
* of selection
[+]preset
[-]track main chain
* create a new object which tracks the
* main chain atoms and shows main chain
* and side chain polar contacts
[-]symmetry surface
* create a symmetry view of the selected
* atoms showing the contact surface as well
* as a selection entry of the atoms in
* contact with symmetry mates
* (only includes atoms of the initial selection).
[-]hydrophobic residues
* show hydrophobic residues
* depending on hydrophobic residue scales
* by KandD (Kyte & Doolittle
* J Mol Biol 157:105, 1982)
* Rose (Rose et al.
* Science, 229, 834-838,1985)
* GES (Engelman Engelman et al.
* Annu Rev Biophys Biophys Chem,
* 15, 321-353(1986)
* (no window selection; just raw categories
* are colored by: blue-hydrophile
* green-neutral
* red-hydrophobe )
[-]surface inspection
* create selections for surface
* inspections
# BNI tools additional settings in PyMOL sidebar
[+]color
[-]by ss
* color helix,sheet and loop separately
[.]Helix
[.]Sheet
[.]Loop
# this section is replaced by "[-] by_rep" in PyMOL versions >1.6
[-]surface
* color surface separately from atoms
[.]by atom
* set surface color to standard
[.]by map
* if a map or ramp is loaded
* color surface by ramp/map
[.]my color
* color surface by own defined color
[-]mesh
* color mesh separately from atoms
[.]by atom
* set mesh color to standard
[.]by map
* if a map or ramp is loaded
* color mesh by ramp/map
[.]my color
* color mesh by own defined color
[-]label
* color labels separately
[.]by atom
* set label color by atom
[.]my color
* color labels by own defined color
[-]stick
* color sticks separately
[.]standard
* set stick color to standard
[.]my color
* color sticks by own defined color
[-]my colors
* use/append own defined colors
* own colors can be defined by
* Setting->Colors..->New
* to keep color settings for
* other pymol sessions
* you have to set the colors
* in .pymolrc or similar
* pymol setting file
* like
* set_color mycolor,[ 1.00, 1.00, 1.00]
[+]show
[-]surface flag
* set surface flag of atoms to show hide
* or ignore fro surface calculation
[-]transparency
* set different transparency types
* on selections or atoms