Dssr block: Difference between revisions
(link to blog post) |
PedroLacerda (talk | contribs) No edit summary |
||
| (4 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
{{Infobox script-repo | {{Infobox script-repo | ||
|type = script | |type = script | ||
|filename = dssr_block.py | |filename = script/dssr_block.py | ||
|author = [[User:Speleo3|Thomas Holder]] | |author = [[User:Speleo3|Thomas Holder]] | ||
|license = BSD-2-Clause | |license = BSD-2-Clause | ||
}} | }} | ||
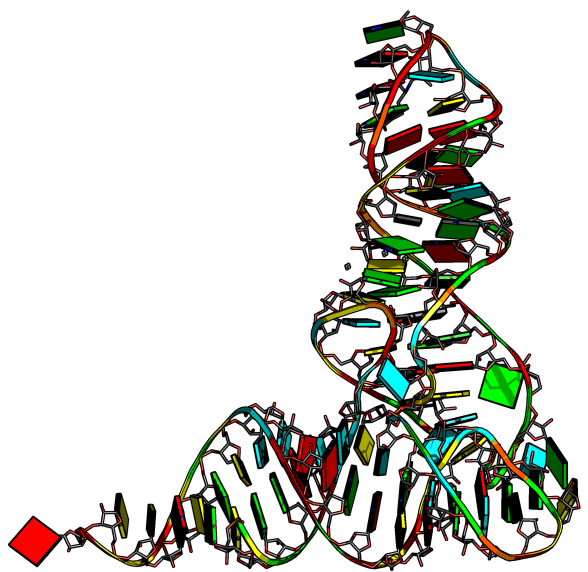
[[File:Trna.png|300px|thumb|right]] | |||
This script adds the [[dssr_block]] command, which is a simple wrapper for the '''DSSR''' program and can create "block" shaped cartoons for nucleic acid bases and base pairs. | This script adds the [[dssr_block]] command, which is a simple wrapper for the '''DSSR''' program and can create "block" shaped cartoons for nucleic acid bases and base pairs. | ||
''Requires the '''x3dna-dssr''' executable, obtainable from http://x3dna.org'' | ''Requires the '''x3dna-dssr''' executable, obtainable from http://x3dna.org'' | ||
Instead of installing '''x3dna-dssr''' and the plugin locally, you can also use the web server at http://skmatic.x3dna.org/ | |||
See also the blog post by DSSR author Xiang-Jun Lu: http://x3dna.org/highlights/dssr-base-blocks-in-pymol-interactively | See also the blog post by DSSR author Xiang-Jun Lu: http://x3dna.org/highlights/dssr-base-blocks-in-pymol-interactively | ||
| Line 24: | Line 28: | ||
dssr_block [ selection [, state [, block_file [, block_depth | dssr_block [ selection [, state [, block_file [, block_depth | ||
[, name [, exe ]]]]]] | [, block_color [, name [, exe ]]]]]]] | ||
== Arguments == | == Arguments == | ||
| Line 32: | Line 36: | ||
* '''block_file''' = face|edge|wc|equal|minor|gray: Corresponds to the '''--block-file''' option (see [http://x3dna.org/ DSSR manual]). Values can be combined, e.g. "wc-minor". {default: face} | * '''block_file''' = face|edge|wc|equal|minor|gray: Corresponds to the '''--block-file''' option (see [http://x3dna.org/ DSSR manual]). Values can be combined, e.g. "wc-minor". {default: face} | ||
* '''block_depth''' = float: thickness of rectangular blocks {default: 0.5} | * '''block_depth''' = float: thickness of rectangular blocks {default: 0.5} | ||
* '''block_color''' = str: Corresponds to the '''--block-color''' option (new in DSSR v1.5.2) {default: } | |||
* '''name''' = str: name of new CGO object {default: dssr_block##} | * '''name''' = str: name of new CGO object {default: dssr_block##} | ||
* '''exe''' = str: path to "x3dna-dssr" executable {default: x3dna-dssr} | * '''exe''' = str: path to "x3dna-dssr" executable {default: x3dna-dssr} | ||
| Line 61: | Line 66: | ||
dssr_block 2n2d, 0 | dssr_block 2n2d, 0 | ||
set all_states | set all_states | ||
</source> | |||
Custom coloring: | |||
<source lang="python"> | |||
fetch 1msy, async=0 | |||
dssr_block block_color=N red | minor 0.9 | major yellow | |||
</source> | </source> | ||
| Line 67: | Line 79: | ||
* [[3DNA]] (old, obsoleted by x3dna-dssr) | * [[3DNA]] (old, obsoleted by x3dna-dssr) | ||
* [[Overview of nucleic acid cartoons]] | * [[Overview of nucleic acid cartoons]] | ||
* http://skmatic.x3dna.org/ | |||
[[Category:Structural_Biology_Scripts]] | [[Category:Structural_Biology_Scripts]] | ||
[[Category:Pymol-script-repo]] | |||
Latest revision as of 23:52, 22 June 2025
| Type | Python Script |
|---|---|
| Download | script/dssr_block.py |
| Author(s) | Thomas Holder |
| License | BSD-2-Clause |
| This code has been put under version control in the project Pymol-script-repo | |
This script adds the dssr_block command, which is a simple wrapper for the DSSR program and can create "block" shaped cartoons for nucleic acid bases and base pairs.
Requires the x3dna-dssr executable, obtainable from http://x3dna.org
Instead of installing x3dna-dssr and the plugin locally, you can also use the web server at http://skmatic.x3dna.org/
See also the blog post by DSSR author Xiang-Jun Lu: http://x3dna.org/highlights/dssr-base-blocks-in-pymol-interactively
Recommended Setup
Place the x3dna-dssr executable into $PATH. Examples:
- Linux/Mac: /usr/bin/x3dna-dssr
- Windows: C:\Program Files\PyMOL\PyMOL\x3dna-dssr.exe
The script can be run, imported as a Python module, or installed with the Plugin Manager.
Usage
dssr_block [ selection [, state [, block_file [, block_depth
[, block_color [, name [, exe ]]]]]]]
Arguments
- selection = str: atom selection {default: all}
- state = int: object state (0 for all states) {default: -1, current state}
- block_file = face|edge|wc|equal|minor|gray: Corresponds to the --block-file option (see DSSR manual). Values can be combined, e.g. "wc-minor". {default: face}
- block_depth = float: thickness of rectangular blocks {default: 0.5}
- block_color = str: Corresponds to the --block-color option (new in DSSR v1.5.2) {default: }
- name = str: name of new CGO object {default: dssr_block##}
- exe = str: path to "x3dna-dssr" executable {default: x3dna-dssr}
Examples
Combining DSSR block representation with regular PyMOL cartoons:
fetch 1ehz, async=0
as cartoon
set cartoon_ladder_radius, 0.1
set cartoon_ladder_color, gray
set cartoon_nucleic_acid_mode, 1
dssr_block
Joined base-pair blocks (block_file=wc):
fetch 1ehz, async=0
dssr_block block_file=wc
Multi-state Example:
fetch 2n2d, async=0
dssr_block 2n2d, 0
set all_states
Custom coloring:
fetch 1msy, async=0
dssr_block block_color=N red | minor 0.9 | major yellow
See Also
- 3DNA (old, obsoleted by x3dna-dssr)
- Overview of nucleic acid cartoons
- http://skmatic.x3dna.org/