Bondpack: Difference between revisions
No edit summary |
fix download link |
||
| (One intermediate revision by one other user not shown) | |||
| Line 1: | Line 1: | ||
{{Infobox script-repo | {{Infobox script-repo | ||
|type = plugin | |type = plugin | ||
| | |download = https://github.com/rasbt/BondPack | ||
|author = Sebastian Raschka | |author = Sebastian Raschka | ||
|license = GNU GENERAL PUBLIC LICENSE | |license = GNU GENERAL PUBLIC LICENSE | ||
| Line 69: | Line 69: | ||
[[Category:Script_Library]] | [[Category:Script_Library]] | ||
[[Category:Structural_Biology_Scripts]] | [[Category:Structural_Biology_Scripts]] | ||
[[Category:Plugins]] | |||
Latest revision as of 02:14, 6 May 2014
| Type | PyMOL Plugin |
|---|---|
| Download | https://github.com/rasbt/BondPack |
| Author(s) | Sebastian Raschka |
| License | GNU GENERAL PUBLIC LICENSE |
BondPack
[edit]A collection of PyMOL plugins to visualize atomic bonds.
Introduction
[edit]PyMOL is without any doubt a great tool to visualize protein and ligand molecules.
However, drawing interactions between atoms can be often quite cumbersome when done manually.
For the sake of convenience, I developed three plugins for PyMOL that will make our life as protein biologists a little bit easier.
All three PyMOL plugins can be installed and used separately; they don't depend on each other, but rather complement each other.
At the end of this article, you will find brief instructions on how to install plugins in PyMOL - a very quick and simple process.
HydroBond
[edit]HydroBond visualizes all potential polar contacts between protein and ligand molecules within a user-specified distance.
The underlying function is based on the different atom types, such as hydrogen bond acceptor and donor atoms,
and thus it is required to have hydrogen atoms present in the structure.
If your structure file doesn't contain hydrogen atoms already, you can add them directly in PyMOL as shown in the image below.
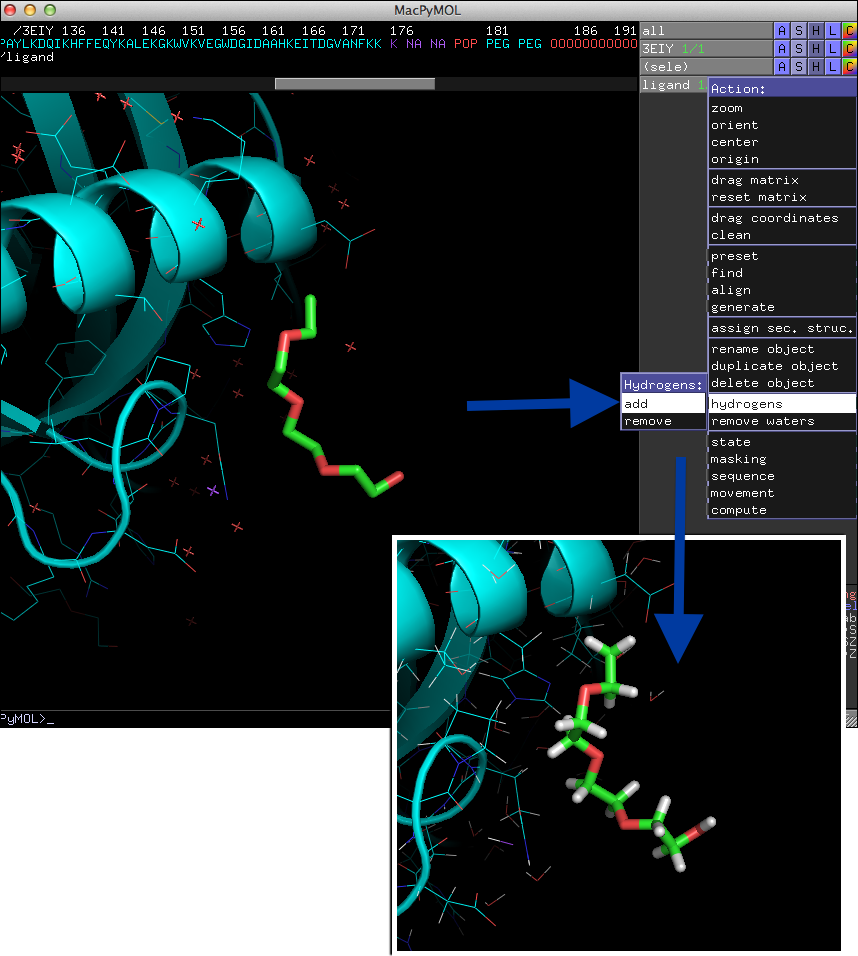
HydroBond is related to PyMOLs "[A]->find->polar contacts" command, however,
it doesn't consider geometry criteria and angle thresholds,
but is rather based on atom types.
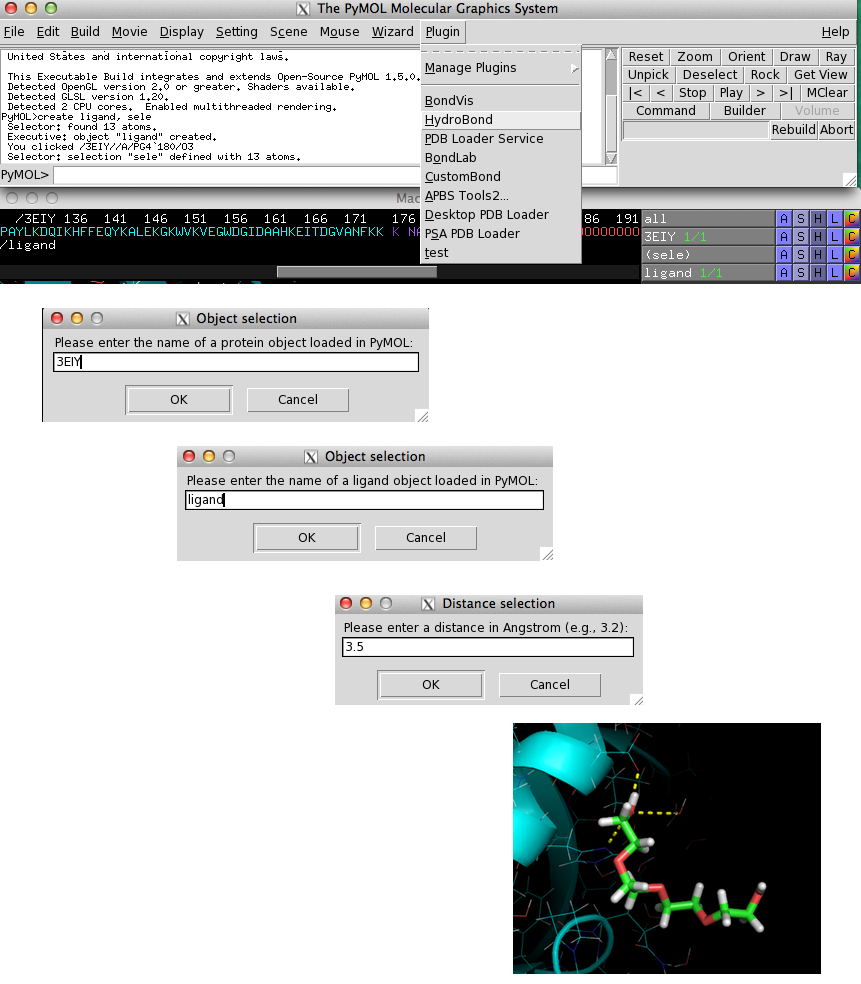
When you select HydroBond from the "Plugin" menu, you will be prompted to enter the name of the protein object,
the ligand object, and a distance cutoff as shown in the figure below.
If HydroBond was able to detect hydrogen bond and acceptor atoms within the
specified distance, potential interactions will be visualized as yellow dashed lines.
BondVis
[edit]The BondVis plugin lets you visualize interactions between any pair of atoms you specified.
Often I find it helpful for my understanding (and for verification) to visualize the bonds between certain atoms
that were assigned in docking or any other prediction software.
Most software will provide you with information about the atoms that were "connected" to perform the analysis.
If you run BondVis from the "Plugin" menu, it will ask you to select a "bond info file."
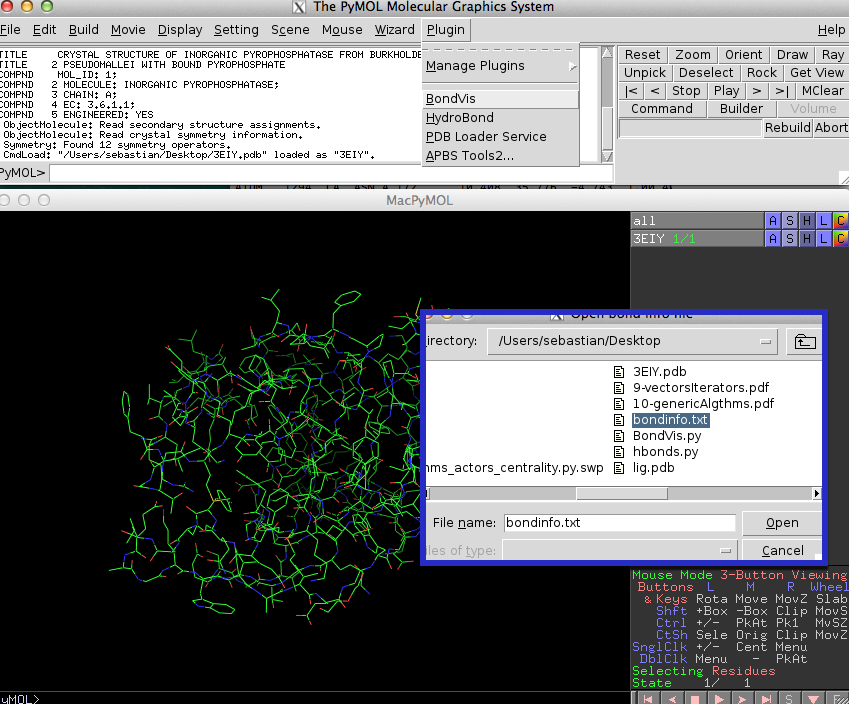
This is just a simple text file that contains the atom numbers of connected atoms in pairs.
Those can be separated by spaces, tabs, or commas. An example file with bond information could look like this:
1174 1357
1175 1358
1176 1359
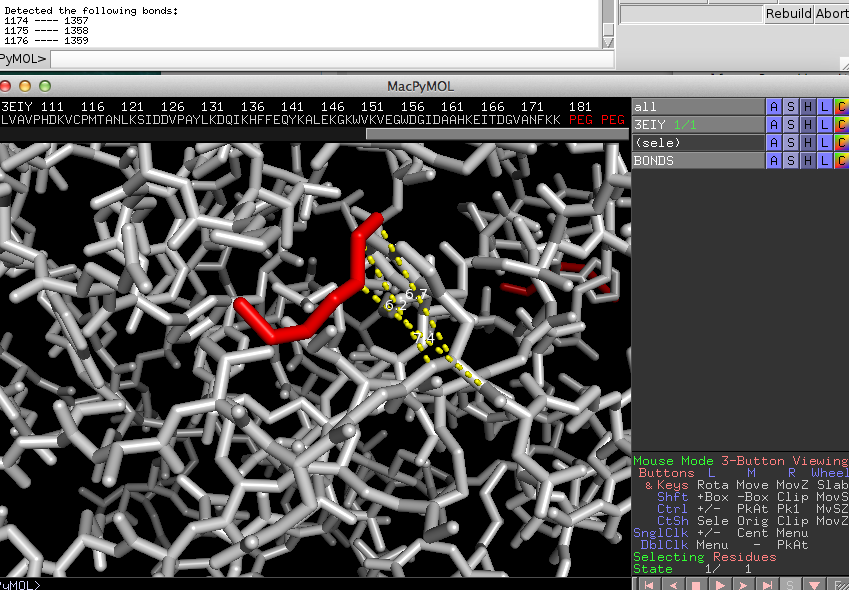
When you selected a "bond info" file, BondVis will connect all atom pairs by yellow dashed lines
and print out the connected atoms in the command field.
BondLab
[edit]If you are not happy with the looks of the lines that were drawn to visualize connections between atoms,
you will like to use BondLab.
This plugin offers a simple interface that allows you to change the diameter,
gap width, and color of the lines.
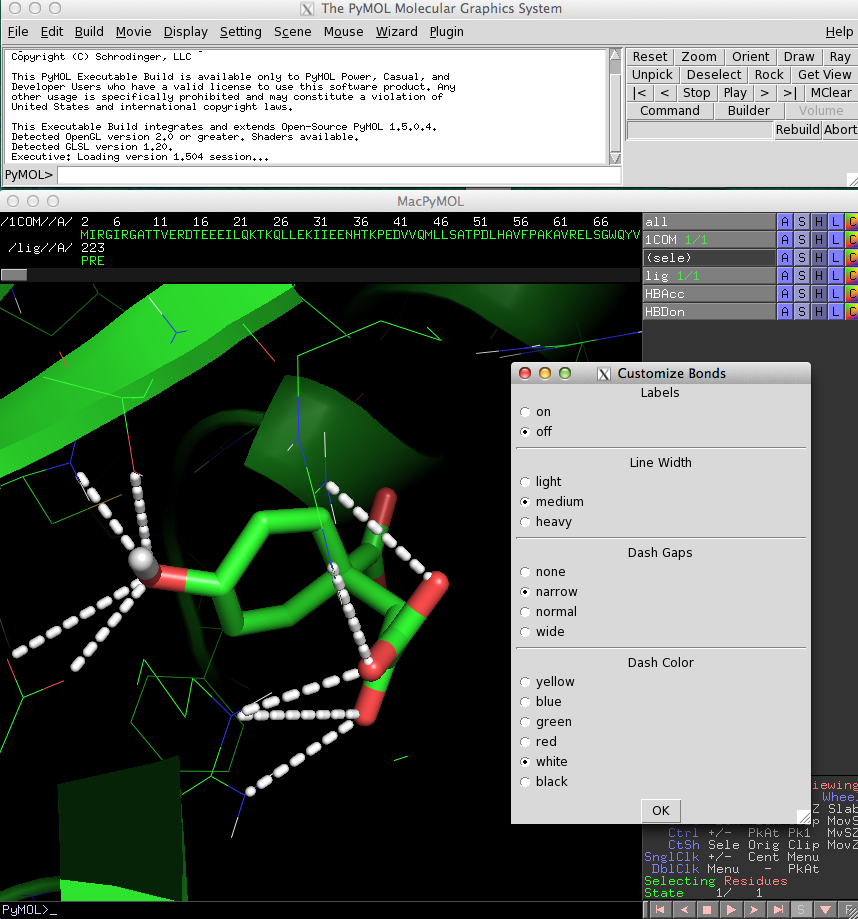
The following video demonstrates the different features of BondLab:
http://youtu.be/14UZctxtK3w
Github
[edit]If you are interested, you can follow the BondPack Github repository
https://github.com/rasbt/BondPack for updates